Chapter 15 - Simulation
15.1 It Is Proven
Unlike statistical research, which is completely made up of things that are at least slightly false, statistics itself is almost entirely true. Statistical methods are generally developed by mathematical proof. If we assume A, B, and C, then based on the laws of statistics we can prove that D is true.
For example, if we assume that the true model is \(Y = \beta_0 + \beta_1X + \varepsilon\), and that \(X\) is unrelated to \(\varepsilon\),398 And some other less commonly-discussed assumptions. then we can prove mathematically that the ordinary least squares estimate of \(\hat{\beta}_1\) will be the true \(\beta_1\) on average. That’s just true, as true as the proof that there are an infinite number of prime numbers. Unlike the infinite-primes case, we can (and do) argue endlessly about whether those underlying true-model and unrelated-to-\(\varepsilon\) assumptions apply in our research case. But we all agree that if they did apply, then the ordinary least squares estimate of \(\hat{\beta}_1\) will be the true \(\beta_1\) on average. It’s been proven.
Pretty much all of the methods and conclusions I discuss in this book are supported by mathematical proofs like this. I have chosen to omit the actual proofs themselves,399 Whether this is a blessing or a deprivation to the reader is up to you (or perhaps your professor). In my defense, there are already plenty of proof-based econometrics books out there for you to peruse. The most famous of which at the undergraduate level is by Wooldridge (2016Wooldridge, Jeffrey M. 2016. Introductory Econometrics: A Modern Approach. Nelson Education.), which is definitely worth a look. Those proof-based books are valuable, but I’m not sure how badly we need another one. Declining marginal value, comparative advantage, and all that. I’m still an economist, you know. but they’re lurking between the lines, and in the citations I can only imagine you’re breezing past.
Why go through the trouble of doing mathematical proofs about our statistical methods? Because they let us know how our estimates work. The proofs underlying ordinary least squares tell us how its sampling variation works, which assumptions must be true for our estimate to identify the causal effect, what will happen if those assumptions aren’t correct, and so on. That’s super important!
So we have an obvious problem. I haven’t taught you how to do econometric proofs. There’s a good chance that you’re not going to go out and learn them, or at least you’re unlikely to go get good at them, even if you do econometric research. Proof-writing is really a task for the people developing methods, not the consumers of those methods. Many (most?) of the people doing active statistical research have forgotten all but the most basic of statistical proofs, if they ever knew them in the first place.400 Whether this is a problem is a matter of opinion. I think doing proofs is highly illuminating, and getting good at doing econometric proofs has made me understand how data works much better. But is it necessary to know the proof behind a method in order to use that method well as a tool? I’m skeptical.
But we still must know how our methods work! Enter simulation. In the context of this chapter, simulation refers to the process of using a random process that we have control over (usually, a data generating process) to produce data that we can evaluate with a given method. Then, we do that over and over and over again.401 And then a few more times. Then, we look at the results over all of our repetitions.
Why do we do this? By using a random process that we have control over, we can choose the truth. That is, we don’t need to worry or wonder about which assumptions apply in our case. We can make assumptions apply, or not apply, by just varying our data generating process. We can choose whether there’s a back door, or a collider, or whether the error terms are correlated, or whether there’s common support.
Then, when we see the results that we get, we can see what kind of results the method will produce given that particular truth. We know the true causal effect since we chose the true causal effect - does our method give us the truth on average? How big is the sampling variation? Is the estimate more or less precise than another method? If we do get the true causal effect, what happens if we rerun the simulation but make the error terms correlated? Will it still give the true causal effect?
It’s a great idea, when applying a new method, or applying a method you know in a context where you’re not sure if it will work, to just go ahead and try it out with a simulation. This chapter will give you the tools you need to do that.
15.1.1 The Anatomy of a Simulation
To give a basic example, imagine we have a coin that we’re going to flip 100 times and then estimate the probability of coming up heads. What estimator might we use? We’d probably just calculate the proportion of heads we get in our sample of \(N = 100\).
What would a simulation look like?
- Decide what our random process/data generating process is. We have control! Let’s say we choose to have a coin that’s truly heads 75% of the time
- Use a random number generator to produce our sample of 100 coin flips, where each flip has a 75% chance of being heads
- Apply our estimator to our sample of 100 coin flips. In other words, calculate the proportion of heads in that sample
- Store our estimate somewhere
- Iterate steps 2-4 a whole bunch of times, drawing a new sample of 100 flips each time, applying our estimator to that sample, and storing the estimate (and perhaps some other information from the estimation process, if we like)
- Look at the distribution of estimates across each of our iterations
That’s it! There’s a lot we can do with the distribution of estimates. First, we might look at the mean of our distribution. The truth that we decided on was 75% heads. So if our estimate isn’t close to 75% heads, that suggests our estimate isn’t very good.
We could use simulation to rule out bad estimators. For example, maybe instead of taking the proportion of heads in the sample to estimate the probability of heads, we take the squared proportion of heads in the sample for some reason. You can try it yourself - applying this estimator to each iteration won’t get us an average estimate of 75% heads.
We can also use this to prove the value of good estimators to ourselves. Don’t take your stuffy old statistics textbook’s word that the proportion of heads is a good estimate of the probability of heads. Try it yourself! You probably already knew your textbook was right. But I find that doing the simulation can also help you to feel that they’re right and understand why the estimator works, especially if you play around with it and try a few variations.402 It’s harder to use simulation to prove the value of a good estimator to others. That’s one facet of a broader downside of simulation - we can only run so many simulations, so we can only show it works for the particular data generating processes we’ve chosen. Proofs can tell you much more broadly and concretely when or where something works or doesn’t.
On a personal note, simulation is far and away the first tool that I reach for when I want to see if an approach I’d like to take makes sense. Got an idea for a method, or a tweak to an existing method, or just a way you’d like to transform or incorporate a particular variable? Try simulating it and see whether the thing you want to do works. It’s not too difficult, and it’s certainly easier than not checking, finishing your project, and finding out at the end that it didn’t work when people read it and tell you how wrong you are.
Aside from looking at whether our estimator can nail the true value, we can also use simulation to understand sampling variation. Sure, on average we get 75% heads, but what proportion of samples will get 50% heads? Or 90%? What proportion will find the proportion to be statistically significantly different from 75% heads?
Simulation is an important tool for understanding an estimator’s sampling distribution. The sampling distribution (and the standard errors it gives us that we want so badly) can be really tricky to figure out, especially if analysis has multiple steps in it, or uses multiple estimators. Simulation can help to figure out the degree of sampling variation without having to write a proof.
Sure, we don’t need simulation to understand the standard error of our coin-flip estimator. The answer is written out pretty plainly on Wikipedia if you head for the right page. But what’s the standard error if we first use matching to link our coin to other coins based on its observable characteristics, apply a smoothing function to account for surprising coin results observed in the past, and then drop missing observations from when we forgot to write down some of the coin flips? Uhhh… I’m lost. I really don’t want to try to write a proof for that standard error. And if someone has already written it for me, I’m not sure I’d be able to find it and understand it.403 Or, similarly, what if the complexity is in the data rather than the estimator? How does having multiple observations per person affect the standard error? Or one group nested within another? Or what if treatments are given to different groups at different times, does that affect standard errors? Simulation can help us get a sense of sampling variation even in complex cases.
Indeed, even the pros who know how to write proofs often use simulation for sampling variation - see the bootstrap standard errors and power analysis sections in this chapter.
Lastly, simulation is crucial in understanding how robust an estimation method can be to violations of its assumptions, and how sensitive its estimates are to changes in the truth. Sure, we know that if a true model has a second-order polynomial for \(X\), but we estimate a linear regression without that polynomial, the results will be wrong. But how wrong will it be? A little wrong? A lot wrong? Will it be more wrong when \(X\) is a super strong predictor or a weak one? Will it be more wrong when the slope is positive or negative, or does it even matter?
It’s very common, when doing simulations, to run multiple variations of the same simulation. Tweak little things about the assumptions - make \(X\) a stronger predictor in one and a weaker predictor in the other. Add correlation in the error terms. Try stuff! You’ve got a toy; now it’s your job to break it so you know exactly when and how it breaks.
15.1.2 Why We Simulate
To sum up, simulation is the process of using a random process that we control to repeatedly apply our estimator to different samples. This lets us do a few very important things:
- We can test whether our estimate will, when applied to the random process / data generating process we choose, give us the true effect
- We can get a sense of the sampling distribution of our estimate
- We can see how sensitive an estimator is to certain assumptions. If we make those assumptions no longer true, does the estimator stop working?
Now that we know why simulation can be helpful,404 And I promise you it is. Once you get the hang of it you’ll be doing simulations all the time! They’re not just useful, they’re addictive. let’s figure out how to do it.
15.2 Alas, We Must Code
There’s no way around it. Modern simulation is about using code. And you won’t just have to use prepackaged commands correctly - unless you’re willing to trust an AI to write code this detail-oriented without missing something, you’ll actually have to write your own.405 On one hand, sorry. On the other hand, if this isn’t something you’ve learned to do yet, it is one thousand million billion percent worth your time to figure out, even if you never run another simulation again in your life. As I write this, it’s the 2020s. Computers run everything around you, and are probably the single most powerful type of object you interact with on a regular basis. Don’t you want to understand how to tell those objects what to do? I’m not sure how to effectively get across to you that even if you’re not that good at it, learning how to program a little bit, and how to use a computer well, is actual, tangible power in this day and age. That must be appealing. Right?
Thankfully, not only are simulations relatively straightforward, they’re also a good entryway into writing more complicated programs.
A typical statistical simulation has only a few parts:
- A process for generating random data
- An estimation
- A way of iterating steps 1-2 many times
- A way of storing the estimation results from each iteration
- A way of looking at the distribution of estimates across iterations
Let’s walk through the processes for coding each of these.
How can we code a data generating process? This is a key question. Often, when you’re doing a simulation, the whole point of it will be to see how an estimator performs under a particular set of data generating assumptions. So we need to know how to make data that satisfies those assumptions.
The first question is how to produce random data in general.406 If you want to get picky, this chapter is all about pseudo-random data. Most random-number generators in programming languages are actually quite predictable since, y’know, computers are pretty predictable. They start with a “seed.” They run that seed through a function to generate a new number and also a new seed to use for the next number. For our purposes, pseudo-random is fine. Perhaps even better than actually-random, because it means we can set our own seed to make our analysis reproducible, i.e., everyone should get the same result running our code. There are, generally, functions for generating random data according to all sorts of distributions (as described in Chapter 3). But we can often get by with a few:
- Random uniform data, for generating variables that must lie within certain bounds.
runif()in R,runiform()in Stata, andnumpy.random.default_rng().uniform()in Python.407 Importantly, pretty much any random distribution can be generated using random uniform data, by running it through an inverse conditional distribution function. Or for categorical data, you can do things like “if \(X > .6\) then \(A = 2\), if \(X <=. 6\) and \(X >= .25\) then \(A = 1\), and otherwise \(A = 0\),” where \(X\) is uniformly distributed from 0 to 1. This is the equivalent of a creating a categorical \(A\), which is 2 with probability .4 (\(1 - .6\)), 1 with probability .35 (\(.6 - .25\)), and 0 with probability .25. - Normal random data, for generating continuous variables, and in many cases mimicking real-world distributions.
rnorm()in R,rnormal()in Stata, andnumpy.random.default_rng().normal()in Python. - Categorical random data, for categorical or binary variables.
sample()in R,runiformint()in Stata, ornumpy.random.default_rng().choice()in Python. The Stata function will only work if you want an equal chance of each category. Want unequal probabilities in Stata? Try carving up uniform data, as in the previous note. Or if you have only two categories, userbinomial(1,p).
You can use these simple tools to generate the data generating processes you like. For example, take the code snippets a few paragraphs after this in which I generate a data set with a normally distributed \(\varepsilon\) error variable I call \(eps\),408 Yes, we can actually see the error term, usually unobservable! This is the beauty of creating the truth ourselves. This even lets you do stuff like, say, check how closely the residuals from your model match the error term. a binary \(X\) variable which is 1 with probability .2 and 0 otherwise, and a uniform \(Y\) variable.
R Code
library(tidyverse)
# If we want the random data to be the same every time, set a seed
set.seed(1000)
# tibble() creates a tibble; it's like a data.frame
# We must pick a number of observations, here 200
d <- tibble(eps = rnorm(200), # by default mean 0, sd 1
Y = runif(200), # by default from 0 to 1
X = sample(0:1, # sample from values 0 and 1
200, # get 200 observations
replace = TRUE, # sample with replacement
prob = c(.8, .2))) # Be 1 20% of the timeStata Code
* Usually we want a blank slate
clear
* How many observations do we want? Here, 200
set obs 200
* If we want the random data to be the same every time, set a seed
set seed 1000
* Normal data is by default mean 0, sd 1
g eps = rnormal()
* Uniform data is by default from 0 to 1
g Y = runiform()
* For unequal probabilities with binary data,
* use rbinomial
g X = rbinomial(1, .2)Python Code
import pandas as pd
import numpy as np
# Create a reusable generator.
# If we want the results to be the same every time, set a seed
rng = np.random.default_rng(1000)
# Create a DataFrame with pd.DataFrame. The size argument of the random
# functions gives us the number of observations
d = pd.DataFrame({
# normal data is by default mean 0, sd 1
'eps': rng.normal(size = 200),
# Uniform data is by default from 0 to 1
'Y': rng.uniform(size = 200),
# We can use binomial to make binary data
# with unequal probabilities
'X': rng.binomial(1, .2, size = 200)
})Using these three random-number generators,409 And others, if relevant. Read the documentation to learn about others. It’s help(Distributions) in R, help random in Stata, and help(numpy.random) in Python. I keep saying to read the documentation because it’s really good advice! we can then mimic causal diagrams. This is, after all, a causal inference textbook. We want to know how our estimators will fare in the face of different causal relationships!
15.2.1 Creating Data
How can we create data that follows a causal diagram? Easy! If we use variable \(A\) to create variable \(B\), then \(A \rightarrow B\). Simple as that. Take the below code snippets, for example, which modify what we had above to add a back-door confounder \(W\) (and make \(Y\) normal instead of uniform). These code snippets, in other words, are representations of Figure 15.1.
Figure 15.1: Causal Diagram to Be Replicated by Simulation Code
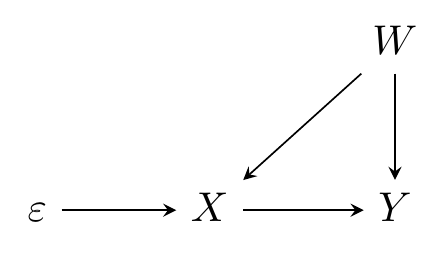
As in Figure 15.1, we have that \(W\) causes \(X\) and \(Y\), so we put in that \(W\) makes the binary \(X\) variable more likely to be 1, but reduces the value of the continuous \(Y\) variable. We also have that \(\varepsilon\) is a part of creating \(X\), since it causes \(X\), and that \(X\) is a part of creating \(Y\), with a true effect of \(X\) on \(Y\) being that a one-unit increase in \(X\) causes a 3-unit increase in \(Y\).410 By the way, if you want to include heterogeneous treatment effects in your code, it’s easy. Just create the individual’s treatment effect like a regular variable. Then use it in place of your single true effect (i.e., here, in place of the 3) when making your outcome. Finally, it’s not listed on the diagram,411 Additional sources of pure randomness are often left off of diagrams because they add a lot of ink and we tend to assume they’re there anyway. but we have an additional source of randomness, \(\nu\), in the creation of \(Y\). If the only arrows heading into \(Y\) were truly \(X\) and \(W\), then \(Y\) would be completely determined by those two variables - no other variation once we control for them.
But presumably there’s something else going into \(Y\), and that’s \(\nu\), which is also the error term if we regress \(Y\) on \(X\) and \(W\). You can imagine \(\nu\) being added to Figure 15.1 with an arrow \(\nu\rightarrow Y\). Where’s \(\nu\) in the code? It’s that random normal data being added in when \(Y\) is created. You’ll also notice that \(eps\) has left the code, too. It’s actually still in there - it’s the uniform random data! We create \(X\) by checking if some uniform random data (\(\varepsilon\)) is lower than .2 plus \(W\) - higher \(W\) values make it more likely we’ll be under \(.2+W\), and so more likely that \(X = 1\). “Carving up” the different values of a uniform random variable can sometimes be easier than working with binomial random data, especially if we want the probability to be different for different observations.
R Code
library(tidyverse)
# If we want the random data to be the same every time, set a seed
set.seed(1000)
# tibble() creates a tibble; it's like a data.frame
# We must pick a number of observations, here 200
d <- tibble(W = runif(200, 0, .1)) %>% # only go from 0 to .1
mutate(X = runif(200) < .2 + W) %>%
mutate(Y = 3*X + W + rnorm(200)) # True effect of X on Y is 3Stata Code
* Usually we want a blank slate
clear
* How many observations do we want? Here, 200
set obs 200
* If we want the random data to be the same every time, set a seed
set seed 1000
* W only varies from 0 to .1
g W = runiform(0, .1)
g X = runiform() < .2 + W
* The true effect of X on Y is 3
g Y = 3*X + W + rnormal()Python Code
import pandas as pd
import numpy as np
# If we want the results to be the same every time, set a seed
rng = np.random.default_rng(1000)
# Create a DataFrame with pd.DataFrame. The size argument of the random
# functions gives us the number of observations
d = pd.DataFrame({
# Have W go from 0 to .1
'W': rng.uniform(0, .1, size = 200)})
# Higher W makes X = 1 more likely
d['X'] = rng.uniform(size = 200) < .2 + d['W']
# The true effect of X on Y is 3
d['Y'] = 3*d['X'] + d['W'] + rng.normal(size = 200)We can go one step further. Not only do we want to randomly create this data, we are going to want to do so a bunch of times as we repeatedly run our simulation. Any time we have code we want to run a bunch of times, it’s often a good idea to put it in its own function, which we can repeatedly call. We’ll also give this function an argument that lets us pick different sample sizes.
R Code
library(tidyverse)
# Make sure the seed goes OUTSIDE the function. It makes the random
# data the same every time, but we want DIFFERENT results each time
# we run it (but the same set of different results, thus the seed)
set.seed(1000)
# Make a function with the function() function. The "N = 200" argument
# gives it an argument N that we'll use for sample size. The "=200" sets
# the default sample size to 200
create_data <- function(N = 200) {
d <- tibble(W = runif(N, 0, .1)) %>%
mutate(X = runif(N) < .2 + W) %>%
mutate(Y = 3*X + W + rnorm(N))
# Use return() to send our created data back
return(d)
}
# Run our function!
create_data(500)Stata Code
* Make sure the seed goes OUTSIDE the function. It makes the random data
* the same every time, but we want DIFFERENT results each time we run it
* (but the same set of different results, thus the seed)
set seed 1000
* Create a function with program define. It will throw an error if there's
* already a function with that name, so we must drop it first if it's there
* But ugh... dropping will error if it's NOT there. So we capture the drop
* to ignore any error in dropping
capture program drop create_data
program define create_data
* `1': first thing typed after the command name, which can be our N
local N = `1'
clear
set obs `N'
g W = runiform(0, .1)
g X = runiform() < .2 + W
* The true effect of X on Y is 3
g Y = 3*X + W + rnormal()
* End ends the program
* Unlike the other languages we aren't returning anything,
* instead the data we've created here is just... our data now.
end
* Run our function!
create_data 500Python Code
import pandas as pd
import numpy as np
# Make sure to create the generator OUTSIDE the function.
# We want DIFFERENT results each time we run the function.
# (but the same set of different results, thus the seed)
rng = np.random.default_rng(1000)
# Make a function with def. The "N = 200" argument gives it an argument N
# that we'll use for sample size. The "=200" sets the default to 200
def create_data(N = 200):
d = pd.DataFrame({
'W': rng.uniform(0, .1, size = N)})
d['X'] = rng.uniform(size = N) < .2 + d['W']
d['Y'] = 3*d['X'] + d['W'] + rng.normal(size = N)
# Use return() to send our created data back
return(d)
# And run our function!
create_data(500)What if we want something more complex? We’ll just have to think more carefully about how we create our data!
A common need in creating more-complex random data is in mimicking a panel structure. How can we create data with multiple observations per individual? Well, we create an individual identifier, we give that identifier some individual characteristics, and then we apply those to the entire time range of data. This is easiest to do by merging an individual-data set with an individual-time data set in R and Python, or by use of egen in Stata.
R Code
library(tidyverse)
set.seed(1000)
# N for number of individuals, T for time periods
create_panel_data <- function(N = 200, T = 10) {
# Create individual IDs with the crossing()
# function, which makes every combination of two vectors
# (if you want some to be incomplete, drop them later)
panel_data <- crossing(ID = 1:N, t = 1:T) %>%
# And individual/time-varying data
# (n() means "the number of rows in this data"):
mutate(W1 = runif(n(), 0, .1))
# Now an individual-specific characteristic
indiv_data <- tibble(ID = 1:N,
W2 = rnorm(N))
# Join them
panel_data <- panel_data %>%
full_join(indiv_data, by = 'ID') %>%
# Create X, caused by W1 and W2
mutate(X = 2*W1 + 1.5*W2 + rnorm(n())) %>%
# And create Y. The true effect of X on Y is 3
# But W1 and W2 have causal effects too
mutate(Y = 3*X + W1 - 2*W2 + rnorm(n()))
return(panel_data)
}
create_panel_data(100, 5)Stata Code
* The first() egen function is in egenmore; ssc install egenmore if necessary
set seed 1000
capture program drop create_panel_data
program define create_panel_data
* `1': the first thing typed after the command name, which can be N
local N = `1'
* The second thing can be our T
local T = `2'
clear
* We want N*T observations
local obs = `N'*`T'
set obs `obs'
* Create individual IDs by repeating 1 T times, then 2 T times, etc.
* We can do this with floor((_n-1)/`T') since _n is the row number
* Think about why this works!
g ID = floor((_n-1)/`T')
* Now we want time IDs by repeating 1, 2, 3, ..., T a full N times.
* We can do this with mod(_n,`T'), which gives the remainder from
* _n/`T'. Think about why this works!
g t = mod((_n-1),`T')
* Individual and time-varying data
g W1 = rnormal()
* This variable will be constant at the individual level.
* We start by making a variable just like W1
g temp = rnormal()
* But then use egen to apply it to all observations with the same ID
by ID, sort: egen W2 = first(temp)
drop temp
* Create X based on W1 and W2
g X = 2*W1 + 1.5*W2 + rnormal()
* The true effect of X on Y is 3 but W1 and W2 cause Y too
g Y = 3*X + W1 - 2*W2 + rnormal()
end
create_panel_data 100 5Python Code
import pandas as pd
import numpy as np
from itertools import product
rng = np.random.default_rng(1000)
# N for number of individuals, T for time periods
def create_panel_data(N = 200, T = 10):
# Use product() to get all combinations of individual and
# time (if you want some to be incomplete, drop later)
p = pd.DataFrame(
product(range(0,N), range(0, T)))
p.columns = ['ID','t']
# Individual- and time-varying variable
p['W1'] = rng.normal(size = N*T)
# Individual data
indiv_data = pd.DataFrame({
'ID': range(0,N),
'W2': rng.normal(size = N)})
# Bring them together
p = pd.merge(p, indiv_data, on = 'ID')
# Create X, caused by W1 and W2
p['X'] = 2*p['W1'] + 1.5*p['W2'] + rng.normal(size = N*T)
# And create Y. The true effect of X on Y is 3
# But W1 and W2 have causal effects too
p['Y'] = 3*p['X'] + p['W1']- 2*p['W2'] + rng.normal(size = N*T)
return(p)
create_panel_data(100, 5)Another pressing concern in the creation of random data is in creating correlated error terms. If we’re worried about the effects of non-independent error terms, then simulation can be a great way to try stuff out, if we can make errors that are exactly as dependent as we like. There are infinite varieties to consider, but I’ll consider two: heteroskedasticity and clustering.
Heteroskedasticity is relatively easy to create. Whenever you’re creating variables that are meant to represent the error term, we just allow the variance of that data to vary based on the observation’s characteristics.
R Code
Stata Code
set seed 1000
capture program drop create_het_data
program define create_het_data
* `1`: the first thing typed after the command name, which can be N
local N = `1'
clear
set obs `N'
g X = runiform()
* Let the standard deviation of the error
* Be related to X. Heteroskedasticity!
g Y = 3*X + rnormal(0, 5*X)
end
create_het_data 500Python Code
import pandas as pd
import numpy as np
rng = np.random.default_rng(1000)
def create_het_data(N = 200):
d = pd.DataFrame({
'X': rng.uniform(size = N)})
# Let the standard deviation of the error
# Be related to X. Heteroskedasticity!
d['Y'] = 3*d['X'] + rng.normal(scale = 5*d['X'])
return(d)
create_het_data(500)If we plot the result of create_het_data we get Figure 15.2. The heteroskedastic errors are pretty apparent! We get much more variation around the regression line on the right side of the graph than we do on the left. This is a classic “fan” shape for heteroskedasticity.
Figure 15.2: Simulated Data with Heteroskedastic Errors
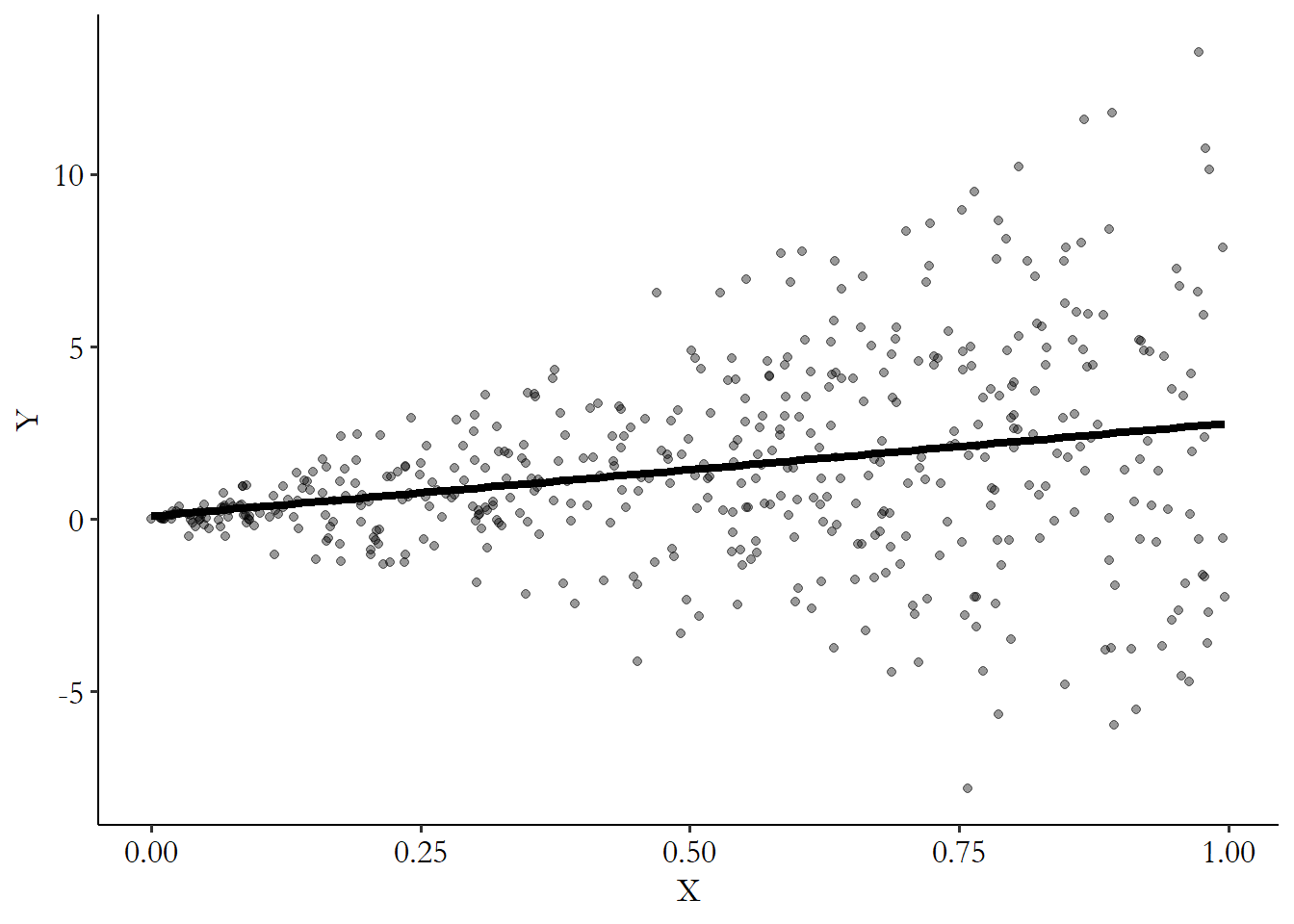
Clustering is just a touch harder. First, we need something to cluster at. This is often some group-identifier variable like the ones we talked about earlier in this section when discussing how to mimic panel structure. Then, we can create clustered standard errors by creating or randomly generating a single “group effect” that can be shared by the group itself, adding on individual noise.
In other words, generate an individual-level error term, then an individual/time-level error term, and add them together in some fashion to get clustered standard errors. Thankfully, we can reuse a lot of the tricks we learned for creating panel data.
R Code
library(tidyverse)
set.seed(1000)
# N for number of individuals, T for time periods
create_clus_data <- function(N = 200, T = 10) {
# We're going to create errors clustered at the
# ID level. So we can follow our steps from making panel data
panel_data <- crossing(ID = 1:N, t = 1:T) %>%
# Individual/time-varying data
mutate(W = runif(n(), 0, .1))
# Now an individual-specific error cluster
indiv_data <- tibble(ID = 1:N,
C = rnorm(N))
# Join them
panel_data <- panel_data %>%
full_join(indiv_data, by = 'ID') %>%
# Create X, caused by W1 and W2
mutate(X = 2*W + rnorm(n())) %>%
# The error term has two components: the individual
# cluster C, and the individual-and-time-varying element
mutate(Y = 3*X + (C + rnorm(n())))
return(panel_data)
}
create_clus_data(100,5)Stata Code
* The first() egen function is in egenmore; ssc install egenmore if necessary
set seed 1000
capture program drop create_clus_data
program define create_clus_data
* `1': the first thing typed after the command name, which can be N
local N = `1'
* The second thing can be our T
local T = `2'
clear
* We want N*T observations
local obs = `N'*`T'
set obs `obs'
* We'll be making errors clustered by ID
* so we can follow the same steps from when we made panel data
g ID = floor((_n-1)/`T')
g t = mod((_n-1),`T')
* Individual and time-varying data
g W = rnormal()
* An error component at the individual level
g temp = rnormal()
* Use egen to apply the value to all observations with the same ID
by ID, sort: egen C = first(temp)
drop temp
* Create X
g X = 2*W + rnormal()
* The error term has two components: the individual cluster C,
* and the individual-and-time-varying element
g Y = 3*X + (C + rnormal())
end
create_clus_data 100 5Python Code
import pandas as pd
import numpy as np
from itertools import product
rng = np.random.default_rng(1000)
# N for number of individuals, T for time periods
def create_clus_data(N = 200, T = 10):
# We're going to create errors clustered at the
# ID level. So we can follow our steps from making panel data
p = pd.DataFrame(
product(range(0,N), range(0, T)))
p.columns = ['ID','t']
# Individual- and time-varying variable
p['W'] = rng.normal(size = N*T)
# Now an individual-specific error cluster
indiv_data = pd.DataFrame({
'ID': range(0,N),
'C': rng.normal(size = N)})
# Bring them together
p = pd.merge(p, indiv_data, on = 'ID')
# Create X
p['X'] = 2*p['W'] + rng.normal(size = N*T)
# And create Y. The error term has two components: the individual
# cluster C, and the individual-and-time-varying element
p['Y'] = 3*p['X'] + (p['C'] + rng.normal(size = N*T))
return(p)
create_clus_data(100, 5)How about our estimator? No causal-inference simulation is complete without an estimator. Code-wise, we know the drill. Just, uh, run the estimator we’re interested in on the data we’ve just randomly generated.
Let’s say we’ve read in our textbook that clustered errors only affect the standard errors of a linear regression, but don’t bias the estimator. We want to confirm that for ourselves. So we take some data with clustered errors baked into it, and we estimate a linear regression on that data.
The tricky part at this point is often figuring out how to get the estimate you want (a coefficient, usually), out of that regression so you can return it and store it. If what you’re going after is a coefficient from a linear regression, you can get the coefficient on the variable varname in R with coef(m)['varname'] for a model m, in Stata with _b[varname] just after running the model, and in Python with m.params['varname'] for a model m.412 To extract something besides a coefficient, you might have to do a bit more digging. But often an Internet search for “how do I get STATISTIC from ANALYSIS in LANGUAGE” will do it for you.
Since this is, again, something we’ll want to call repeatedly, we’re going to make it its own function, too. A function that, itself, calls the data generating function we just made! We’ll be making reference to the create_clus_data function we made in the previous section.
R Code
library(tidyverse)
set.seed(1000)
# A function for estimation
est_model <- function(N, T) {
# Get our data. This uses create_clus_data from earlier
d <- create_clus_data(N, T)
# Run a model that should be unbiased
# if clustered errors themselves don't bias us!
m <- lm(Y ~ X + W, data = d)
# Get the efficient on X, which SHOULD be true value 3 on average
x_coef <- coef(m)['X']
return(x_coef)
}
# Run our model
est_model(200, 5)Stata Code
set seed 1000
capture program drop est_model
program define est_model
* `1: the first thing typed after the command name, which can be N
local N = `1'
* The second thing can be our T
local T = `2'
* Create our data. This uses create_clus_data from earlier
create_clus_data `N' `T'
* Run a model that should be unbiased
* if clustered errors themselves don't bias us!
reg Y X W
end
* Run our model
est_model 200 5
* and get the estimate
display _b[X] Python Code
import pandas as pd
import numpy as np
from itertools import product
import statsmodels.formula.api as smf
rng = np.random.default_rng(1000)
def est_model(N = 200, T = 10):
# This uses create_clus_data from earlier
d = create_clus_data(N, T)
# Run a model that should be unbiased
# if clustered errors themselves don't bias us!
m = smf.ols('Y ~ X + W', data = d).fit()
# Get the coefficient on X, which SHOULD be true value 3 on average
x_coef = m.params['X']
return(x_coef)
# Estimate our model!
est_model(200, 5)Now we must iterate. We can’t just run our simulation once - any result we got might be a fluke. Plus, we usually want to know about the distribution of what our estimator gives us, either to check how spread out it is or to see if its mean is close to the truth. Can’t get a sampling distribution without multiple samples. How many times should you run your simulation? There’s no hard-and-fast number, but it should be a bunch! You can run just a few iterations when testing out your code to make sure it works, but when you want results you should go for more. As a rule of thumb, the more detail you want in your results, or the more variables there are in your estimation, the more you should run - want to see the bootstrap distribution of a mean? A thousand or so should be more than you need. Want a correlation or linear regression coefficient? A few thousand. Want to precisely estimate a bunch of percentiles of a sampling distribution? Maybe go for a number in the tens of thousands.
Iteration is one thing that computers are super good at. In all three of our code examples, we’ll be using a for loop in slightly different ways. A for loop is a computer-programming way of saying “do the same thing a bunch of times, once for each of these values.”
For example, the (generic-language/pseudocode) for loop:
for i in [1, 4, 5, 10]: print i
would do the same thing (print i) a bunch of times, once for each of the values we’ve listed (1, 4, 5, and 10). First, it would do it with i set to 1, and so print the number 1. Then it would do it with i set to 4, and so print the number 4. Then it would print 5, and finally it would print 10.
In the case of simulation, our for loop would look like:
for i in [1 to 100]: do my simulation
where we’re not using the iteration number i at all,413 Except perhaps to store the results - we’ll get there. but instead just repeating our simulation, in this case 100 times.
The way this loop ends up working is slightly different in each language. Python has the purest, “most normal” loop, which will look a lot like the generic language. R could also look like that, but I’m going to recommend the use of the purrr package and its map functions, which will save us a step later. For Stata, the loop itself will be pretty normal, but the fact that we’re creating new data all the time, plus Stata’s limitation of only using one data set at a time, is going to require us to clear our data out over and over (plus, it will add a step later).
All of these code examples will refer back to the create_clus_data and est_model functions we defined in earlier code chunks.
R Code
library(tidyverse)
library(purrr)
set.seed(1000)
# Estimate our model 1000 times (from 1 to 1000)
estimates <- 1:1000 %>%
# Run the est_model function each time
map_dbl(function(x) est_model(N = 200, T = 5))
# There are many map functions in purrr. Since est_model outputs a
# number (a "double"), we can use map_dbl and get a vector of estimatesStata Code
Python Code
import pandas as pd
import numpy as np
from itertools import product
import statsmodels.formula.api as smf
rng = np.random.default_rng(1000)
# This runs est_model once for each iteration as it iterates through
# the range from 0 to 999 (1000 times total)
estimates = [est_model(200, 5) for i in range(0,1000)]Now for a way of storing the results. We already have a function that runs our estimation, which itself calls the function that creates our simulated data. We need a way to store those estimation results.
In both R and Python, our approach to iteration is enough. The map() functions in R, and the standard for-loop structure in Python, give us back a vector of results over all the iterations. So for those languages, we’re already done. The last code chunk already has the instructions you need.
Stata is a bit trickier given its preference for only having one data set at a time. How can you store the randomly generated data and the result-of-estimation data at once? We can use the preserve/restore trick - store the results in a data set, preserve that data set so Stata remembers it, then clear everything and run our estimation, save the estimate as a local, restore back to our results data set, and assign that local into data to remember it.414 Stata 16 has ways of using more than one data set at once, but I don’t want to assume you have Stata 16, and also I never learned this functionality myself, and suspect most Stata users out there are just preserve/restoring to their heart’s content. We could also try storing the results in a matrix, but if you can get away with avoiding Stata matrices you’ll live a happier life.
Now, I will say, there is a way around this sort of juggling, and that’s to use the simulate command in Stata. With simulate, you specify a single function for creating data and estimating a model, and then pulling all the results back out. help simulate should be all you need to know. I find it a bit less flexible than coding things explicitly and not much less work, so I rarely use it in this book.415 And heck, I’ve been running simulations in Stata since like 2008 and I managed to get by without it until now. But you might want to check it out - you may find yourself preferring it.
Stata Code
set seed 1000
* Our "parent" data set will store our results
clear
* We're going to run 1000 times and so need 1000
* observations to store data in
set obs 1000
* and a variable to store them in
g estimates = .
* Estimate the model 1000 times
forvalues i = 1(1)1000 {
* preserve to store our parent data
preserve
quietly est_model 200 5
* restore to get parent data back
restore
* and save the result in the ith row of data
replace estimates = _b[X] in `i'
}Finally, we evaluate. We can now check whatever characteristics of the sampling estimator we want! With our set of results across all the randomly-generated samples in hand, we can, say, look and see whether the mean of the sampling variation is the true effect we decided.
Let’s check. We know from our data generation function that the true effect of \(X\) on \(Y\) is 3. Do we get an average near 3 in our simulated analysis? A simple mean() in R, summarize or mean in Stata, or np.mean() in Python, gives us a mean of 3.00047. Pretty close! Seems like our textbook was right: clustered errors alone won’t bias an otherwise unbiased estimate.
How about the standard errors? The distribution of estimates across all our random samples should mimic sampling distribution. So, the standard deviation of our results is the standard error of the estimate. sd() in R, summarize in Stata, or np.std() in Python each show a standard deviation of .045.
So an honest linear regression estimator should give standard errors of .045. Does it? We can use simulation to answer that. We can take the simulation code we already have but swap out the part where we store a coefficient estimate to instead store the standard error. This can be done easily with tidy() from the broom package in R,416 If you prefer, you could skip broom and get the standard errors with coef(summary(m))['X', 'Std. Error']. But broom makes extracting so many other parts of the regression object so much easier that I recommend getting used to it. _se[X] in Stata, or m.bse['X'] in Python, where X is the name of the variable you want the standard errors of. When we simulate that a few hundred times, the mean reported standard error is .044. Just a hair too small. If we instead estimate heteroskedasticity-robust standard errors, we get .044. Not any better! If we treat the standard errors as is most appropriate, given the clustered errors we know we created, and use standard errors clustered at the group level, we get .045. Seems like that worked pretty well!
15.3 The Old Man and the C
As I mentioned,417 None of the code examples in this book use the C language, but I hope you’ll excuse me. one of the most powerful things that simulation can do in the context of causal inference is let you know if a given estimator works to provide, on average, the true causal effect. Bake a true causal effect into your data generating function, then put the estimator you’re curious about in the estimation function, and let ’er rip! If the mean of the estimates is near the truth and the distribution isn’t too wide, you’re good to go.
This is a fantastic way to test out any new method or statistical claim you’re exposed to. We already checked whether OLS still works on average with clustered errors. But we could do plenty more. Can one back door cancel another out if the effects on the treatment have opposite signs? Does matching really perform better than regression when one of the controls has a nonlinear relationship with the outcome? Does controlling for a collider really add bias? Make a data set with a back-door path blocked by a collider, and see what happens if you control or don’t control for it in your estimation function.418 By the way, making data with a collider in it is a great way to intuitively convince yourself that their uncontrolled presence doesn’t bias you. To make data with, for example, a collider \(C\) with the path \(X \rightarrow C \leftarrow Y\), you have to first make \(X\), and then make \(Y\) with whatever effect of \(X\), and then finally make \(C\) using both \(X\) and \(Y\). You could have stopped before even making \(C\) and still run a regression of \(Y\) on \(X\). Surely just creating the variable doesn’t change the coefficient.
This makes simulation a great tool for three things: (1) trying out new estimators, (2) comparing different estimators, and (3) seeing what we need to break in order to make an estimator stop working.
We can do all of these things without having to write econometric proofs. Sure, our results won’t generalize as well as a proof would, but it’s a good first step. Plus, it puts the power in our hands!
We can imagine an old man stuck on a desert island somewhere, somehow equipped with a laptop and his favorite statistical software, but without access to textbooks or experts to ask. Maybe his name is Agus. Agus has his own statistical questions - what works? What doesn’t? Maybe he has a neat idea for a new estimator. He can try things out himself. I suggest you do too. Trying stuff out in simulation is not just a good way to get answers on your own, it’s also a good way to get to know how data works.
15.3.1 Trying out New Estimators
Professors tend to show you the estimators that work well. But that can give the false illusion that these estimators fell from above, all of them working properly. Untrue! Someone had to think them up first, try them out, and prove that they worked.
There’s no reason you can’t come up with your own estimator and try it out. Sure, it probably won’t work. But I bet you’ll learn something.
Let’s try our own estimator. Or Agus’ estimator. Agus had a thought: you know how the standard error of a regression gets smaller as the variance of \(X\) gets bigger? Well… what if we only pick the extreme values of \(X\)? That will make the variance of \(X\) get very big. Maybe we’ll end up with smaller standard errors as a result.
So that’s Agus’ estimator: instead of regressing \(Y\) on \(X\) as normal, first limit the data to just observations with extreme values of \(X\) (maybe the top and bottom 20%), and then regress \(Y\) on \(X\) using that data.419 You might be anticipating at this point that this isn’t going to work well. At the very least, if it did I’d probably have already mentioned it in this book or something, right? But do you know why it won’t work well? Just sit and think about it for a while. Then, once you’ve got an idea of why you think it might be… you can try that idea out in simulation! How would you construct a simulation to test whether your explanation of why it doesn’t work is the right one? We think it might reduce standard errors, although we’d also want to make sure it doesn’t introduce bias.
Agus has a lot of ideas like this. He writes them on a palm leaf and then programs them on his old sandy computer to see if they work. For the ones that don’t work, he takes the palm leaf and floats it gently on the ocean, giving it a push so that it drifts out to the horizon, bidden farewell. Will this estimator join the others at sea?
Let’s find out by coding it up ourselves. Can you really code up your own estimator? Sure! Just write code that tells the computer to do what your estimator does. And if you don’t know how to write that code, search the Internet until you do.
There’s a tweak in this code, as well. We’re interested in whether the effect is unbiased, but also whether standard errors will be reduced. So we may want to collect both the coefficient and its standard error. There are two things to return. How can we do it? See the code.
R Code
library(tidyverse); library(purrr); library(broom)
set.seed(1000)
# Data creation function. Let's make the function more flexible
# so we can choose our own true effect!
create_data <- function(N, true) {
d <- tibble(X = rnorm(N)) %>%
mutate(Y = true*X + rnorm(n()))
return(d)
}
# Estimation function. keep is the portion of data in each tail
# to keep. So .2 would keep the bottom and top 20% of X
est_model <- function(N, keep, true) {
d <- create_data(N, true)
# Agus' estimator!
m <- lm(Y~X, data = d %>%
filter(X <= quantile(X, keep) | X >= quantile(X, (1-keep))))
# Return coefficient and standard error as two elements of a list
ret <- tidy(m)
return(list('coef' = ret$estimate[2],
'se' = ret$std.error[2]))
}
# Run 1000 simulations. use map_df to stack all the results
# together in a data frame
results <- 1:1000 %>%
map_df(function(x) est_model(N = 1000,
keep = .2,
true = 2))
mean(results$coef); sd(results$coef); mean(results$se)Stata Code
set seed 1000
* Data creation function
capture program drop create_data
program define create_data
clear
local N = `1'
* The second argument will let us be a bit more flexible -
* we can pick our own true effect!
local true = `2'
set obs `N'
g X = rnormal()
g Y = `true'*X + rnormal()
end
* Model estimation function
capture program drop est_model
program define est_model
local N = `1'
* Our second argument will be the proportion to keep in
* each tail so .2 would mean keep the bottom 20% and top 20%
local keep = `2'
local true = `3'
create_data `N' `true'
* Since X has no missings, if we sort by X, the top and bottom X%
* of X will be in the first and last X% of observations
* Agus' estimator!
sort X
drop if _n > `keep'*`N' & _n < (1-`keep')*`N'
reg Y X
end
* To return multiple things, we need only to have
* Multiple "parent" variables to store them in
clear
set obs 1000
g estimate = .
g se = .
forvalues i = 1(1)1000 {
preserve
quietly est_model 1000 .2 2
restore
replace estimate = _b[X] in `i'
replace se = _se[X] in `i'
}
summ estimate sePython Code
import pandas as pd
import numpy as np
import statsmodels.formula.api as smf
rng = np.random.default_rng(1000)
# Data creation function. Let's also make the function more
# flexible - we can choose our own true effect!
def create_data(N, true):
d = pd.DataFrame({'X': rng.normal(size = N)})
d['Y'] = true*d['X'] + rng.normal(size = N)
return(d)
# Estimation function. keep is the portion of data in each tail
# to keep. So .2 would keep the bottom and top 20% of X
def est_model(N, keep, true):
d = create_data(N, true)
# Agus' estimator!
d = d.loc[(d['X'] <= np.quantile(d['X'], keep)) |
(d['X'] >= np.quantile(d['X'], 1-keep))]
m = smf.ols('Y~X', data = d).fit()
# Return the two things we want as an array
ret = [m.params['X'], m.bse['X']]
return(ret)
# Estimate the results 1000 times
results = [est_model(1000, .2, 2) for i in range(0,1000)]
# Turn into a DataFrame
results = pd.DataFrame(results, columns = ['coef', 'se'])
# Let's see what we got!
(np.mean(results['coef']), np.std(results['coef']),
np.mean(results['se']))If we run this, we see an average coefficient of 2.00 (where the truth was 2), a standard deviation of the coefficient of .0332, and a mean standard error of .0339. See, told you the standard deviation of the simulated coefficient matches the standard error of the coefficient!
So the average is 2.00 relative to the truth of also 2. No bias! Pretty good, right? Agus should be celebrating. Of course, we’re not done yet. We didn’t just want no bias, we wanted lower standard errors than the regular estimation method. How can we get that?
15.3.2 Comparing Estimators
One thing that simulation allows us to do is compare the performance of different methods. You can take the exact same randomly generated data, apply two different methods to it, and see which performs better. This process is often known as a “horse race.”
The horse race method can be really useful when considering multiple different ways of doing the same thing. For example, in Chapter 14 on matching, I mentioned a bunch of different choices that could be made in the matching process that, made one way, would reduce bias, but made another way would improve precision. Allowing a wider caliper brings in more, but worse matches, increasing bias but also making the sampling distribution narrower so you’re less likely to be way off. That’s a tough call to make - lower average bias, or narrower sampling distributions? But it might be easier if it just happens to turn out that the amount of bias reduction you get isn’t very high at all, but the precision savings are massive! How could you know such a thing? Try a horse race simulation.420 Then, of course, the amount of bias and precision you’re trading off would be sensitive to what your data looks like. So you might want to run that simulation with your actual data. You wouldn’t know the “true” value in that case, but you could check balance to get an idea of what bias is. See the bootstrap section below for an idea of how simulation with actual data works.
In the case of Agus’ estimator, we want to know whether it produces lower standard errors than a regular regression. That was the whole idea, right? So let’s check. These code examples all use the create_data function from the previous set of code chunks.
R Code
library(tidyverse); library(purrr);
library(broom); library(vtable)
set.seed(1000)
# Estimation function. keep is the portion of data in each tail
# to keep. So .2 would keep the bottom and top 20% of X
est_model <- function(N, keep, true) {
d <- create_data(N, true)
# Regular estimator!
m1 <- lm(Y~X, data = d)
# Agus' estimator!
m2 <- lm(Y~X, data = d %>%
filter(X <= quantile(X, keep) | X >= quantile(X, (1-keep))))
# Return coefficients as a list
return(list('coef_reg' = coef(m1)[2],
'coef_agus' = coef(m2)[2]))
}
# Run 1000 simulations. Use map_df to stack all the
# results together in a data frame
results <- 1:1000 %>%
map_df(function(x) est_model(N = 1000,
keep = .2, true = 2))
sumtable(results)Stata Code
set seed 1000
* Stata can make horse races a bit tricky since
* _b and _se only report the *most recent* regression results
* But we can store and restore our estimates to get around this!
* Model estimation function
capture program drop est_model
program define est_model
local N = `1'
local keep = `2'
local true = `3'
create_data `N' `true'
* Regular estimator!
reg Y X
estimates store regular
* Agus' estimator!
sort X
drop if _n > `keep'*`N' & _n < (1-`keep')*`N'
reg Y X
estimates store agus
end
* Now for the parent data and our loop
clear
set obs 1000
g estimate_agus = .
g estimate_reg = .
forvalues i = 1(1)1000 {
preserve
quietly est_model 1000 .2 2
restore
* Bring back the Agus model
estimates restore agus
replace estimate_agus = _b[X] in `i'
* And now the regular one
estimates restore regular
replace estimate_reg = _b[X] in `i'
}
* Summarize our results
summ estimate_agus estimate_regPython Code
import pandas as pd
import numpy as np
import statsmodels.formula.api as smf
rng = np.random.default_rng(1000)
def est_model(N, keep, true):
d = create_data(N, true)
# Regular estimator
m1 = smf.ols('Y~X', data = d).fit()
# Agus' estimator!
d = d.loc[(d['X'] <= np.quantile(d['X'], keep)) |
(d['X'] >= np.quantile(d['X'], 1-keep))]
m2 = smf.ols('Y~X', data = d).fit()
# Return the two things we want as an array
ret = [m1.params['X'], m2.params['X']]
return(ret)
# Estimate the results 1000 times
results = [est_model(1000, .2, 2) for i in range(0,1000)]
# Turn into a DataFrame
results = pd.DataFrame(results, columns = ['coef_reg', 'coef_agus'])
# Let's see what we got!
results.describe()What do we get? While both estimators get 2.00 on average for the coefficient, the standard errors are actually lower for the regular estimator. The standard deviation of the coefficient’s distribution across all the simulated samples is .034 for Agus and .031 for the regular estimator. Regular wins!421 A few reasons why Agus’ estimator isn’t an improvement: first, it tosses out data, reducing the effective sample size. Second, while the variance of \(X\) is in the denominator of the standard error, the OLS residuals are in the numerator! Anything that harms prediction, like picking tail data, will harm precision and increase standard errors. Also, picking tail data is just… noisier. Looks like Agus will set his creation adrift at sea once again.
15.3.3 Breaking Things
Statistical analysis is absolutely full to the brim with all kinds of assumptions. Annoying, really. Here’s the thing, though - these assumptions all need to be true for us to know what our estimators are doing. But maybe there are a few assumptions that could be false and we’d still have a pretty good idea what those estimators are doing.
So… how many assumptions can be wrong and we’ll still get away with it? Simulation can help us figure that out.
For example, if we want a linear regression coefficient on \(X\) to be an unbiased estimate of the causal effect of \(X\), we need to assume that \(X\) is unrelated to the error term. In other words, there are no open back doors.
In the social sciences, this is rarely going to be true. There’s always some little thing we can’t measure or control for that’s unfortunately related to both \(X\) and the outcome. So should we never bother with regression?
No way! Instead, we should ask “how big a problem is this likely to be?” and if it’s only a small problem, then hey, close to right is better than far, even if it’s not all the way there.
Simulation can be used to probe these sorts of issues by making a very flexible data creation function, flexible enough to try different kinds of assumption violations or degrees of those violations. Then, run your simulation with different settings and see how bad the results come out. If they’re pretty close to the truth, even with data that violates an assumption, maybe it’s not so bad!422 Do keep in mind when doing this that the importance of certain assumptions, as with everything, can be sensitive to context. If you find that an assumption violation isn’t that bad in your simulation, maybe try doing the simulation again, this time with the same assumption violation but otherwise very different-looking data. If it’s still fine, that’s a good sign!
Perhaps you design your data generation function to have an argument called error_dist. Program it so that if you set error_dist to "normal" you get normally-distributed error terms. "lognormal" and you get log-normally distributed error terms, and so on for all sorts of distributions. Then, you can simulate a linear regression with all different kinds of error distributions and see whether the estimates are still good.
How could we use simulation in application to a remaining open back door? We can program our data generation function to have settings for the strength of that open back door. If we have \(X \leftarrow W \rightarrow Y\) on our diagram, what’s the effect of \(W\) on \(X\), and what’s the effect of \(W\) on \(Y\)? How strong do those two arrows need to be to give us an unacceptably high level of bias?
Once we figure out how bad the problem would need to be to give us unacceptably high levels of bias,423 Which is a bit subjective, and based on context - a .1% bias in the effect of a medicine on deadly side effects might be huge, but a .1% bias in the effect of a job training program on earnings might not be so huge. you can ask yourself how bad the problem is likely to be in your case - if it’s under that bar, you might be okay (although you’d certainly still want to talk about the problem - and why you think it’s okay - in your writeup).424 This whole approach is a form of “sensitivity analysis” - checking how sensitive results are to violations of assumptions. It’s also a form of partial identification, as discussed in Chapter 11 and especially in Chapter @ref{ch-PartialIdentification}. This sensitivity-analysis approach to thinking about remaining open back doors is in common use, and is used to identify a range of reasonable regression coefficients when you’re not sure about open back doors. For example, see Cinelli and Hazlett (2020Cinelli, Carlos, and Chad Hazlett. 2020. “Making Sense of Sensitivity: Extending Omitted Variable Bias.” Journal of the Royal Statistical Society: Series B (Statistical Methodology) 82 (1): 39–67.).
Let’s code this up! We’re going to simply expand our create_data function to include a variable \(W\) that has an effect on both our treatment \(X\) and our outcome variable \(Y\). We’ll then include settings for the strength of the \(W \rightarrow X\) and \(W\rightarrow Y\) relationships. There are, plenty of other tweaks we could add as well - the strength of \(X\rightarrow Y\), the standard deviations of \(X\) and \(Y\), the addition of other controls, nonlinear relationships, the distribution of the error terms, and so on and so on. But let’s keep it simple for now with our two settings.
Additionally, we’re going to take our iteration/loop step and put that in a function too, since we’re going to want to do this iteration a bunch of times with different settings.
R Code
library(tidyverse); library(purrr)
set.seed(1000)
# Have settings for strength of W -> X and for W -> Y
# These are relative to the standard deviation
# of the random components of X and Y, which are 1 each
# (rnorm() defaults to a standard deviation of 1)
create_data <- function(N, effectWX, effectWY) {
d <- tibble(W = rnorm(N)) %>%
mutate(X = effectWX*W + rnorm(N)) %>%
# True effect is 5
mutate(Y = 5*X + effectWY*W + rnorm(N))
return(d)
}
# Our estimation function
est_model <- function(N, effectWX, effectWY) {
d <- create_data(N, effectWX, effectWY)
# Biased estimator - no W control!
# But how bad is it?
m <- lm(Y~X, data = d)
return(coef(m)['X'])
}
# Iteration function! We'll add an option iters for number of iterations
iterate <- function(N, effectWX, effectWY, iters) {
results <- 1:iters %>%
map_dbl(function(x) {
# Let's add something that lets us keep track
# of how much progress we've made. Print every 100th iteration
if (x %% 100 == 0) {print(x)}
# Run our model and return the result
return(est_model(N, effectWX, effectWY))
})
# We want to know *how biased* it is, so compare to true-effect 5
return(mean(results) - 5)
}
# Now try different settings to see how bias changes!
# Here we'll use a small number of iterations (200) to
# speed things up, but in general bigger is better
iterate(2000, 0, 0, 200) # Should be unbiased
iterate(2000, 0, 1, 200) # Should still be unbiased
iterate(2000, 1, 1, 200) # How much bias?
iterate(2000, .1, .1, 200) # Now?
# Does it make a difference whether the effect
# is stronger on X or Y?
iterate(2000, .5, .1, 200)
iterate(2000, .1, .5, 200)Stata Code
set seed 1000
* Have settings for strength of W -> X and for W -> Y
* These are relative to the standard deviation
* of the random components of X and Y, which are 1 each
* (rnormal() defaults to a standard deviation of 1)
capture program drop create_data
program define create_data
* Our first, second, and third arguments
* are sample size, W -> X, and W -> Y
local N = `1'
local effectWX = `2'
local effectWY = `3'
clear
set obs `N'
g W = rnormal()
g X = `effectWX'*W + rnormal()
* True effect is 5
g Y = 5*X + `effectWY'*W + rnormal()
end
* Our estimation function
capture program drop est_model
program define est_model
local N = `1'
local effectWX = `2'
local effectWY = `3'
create_data `1' `2' `3'
* Biased estimator - no W control! But how bad is it?
reg Y X
end
* Iteration function! Option iters determined number of iterations
capture program drop iterate
program define iterate
local N = `1'
local effectWX = `2'
local effectWY = `3'
local iter = `4'
* We don't need all this output! Use quietly{}
quietly {
clear
set obs `iter'
g estimate = .
forvalues i = 1(1)`iter' {
* Let's keep track of how far along we are
* Print every 100th iteration
if (mod(`i',100) == 0) {
* Noisily to counteract the quietly{}
noisily display `i'
}
preserve
est_model `N' `effectWX' `effectWY'
restore
replace estimate = _b[X] in `i'
}
* We want to know *how biased* it is, so compare to true-effect 5
replace estimate = estimate - 5
}
summ estimate
end
* Now try different settings to see how bias changes!
* Here we'll use a small number of iterations (200) to
* speed things up, but in general bigger is better
* Should be unbiased
iterate 2000 0 0 200
* Should still be unbiased
iterate 2000 0 1 200
* How much bias?
iterate 2000 1 1 200
* Now?
iterate 2000 .1 .1 200
* Does it make a difference whether the effect
* is stronger on X or Y?
iterate 2000 .5 .1 200
iterate 2000 .1 .5 200Python Code
import pandas as pd
import numpy as np
import statsmodels.formula.api as smf
rng = np.random.default_rng(1000)
# Have settings for strength of W -> X and for W -> Y
# These are relative to the standard deviation
# of the random components of X and Y, which are 1 each
# (np.random.normal() defaults to a standard deviation of 1)
def create_data(N, effectWX, effectWY):
d = pd.DataFrame({'W': rng.normal(size = N)})
d['X'] = effectWX*d['W'] + rng.normal(size = N)
# True effect is 5
d['Y'] = 5*d['X'] + effectWY*d['W'] + rng.normal(size = N)
return(d)
# Our estimation function
def est_model(N, effectWX, effectWY):
d = create_data(N, effectWX, effectWY)
# Biased estimator - no W control!
# But how bad is it?
m = smf.ols('Y~X', data = d).fit()
return(m.params['X'])
# Iteration function! Option iters determines number of iterations
def iterate(N, effectWX, effectWY, iters):
results = [est_model(N, effectWX, effectWY) for i in range(0,iters)]
# We want to know *how biased* it is, so compare to true-effect 5
return(np.mean(results) - 5)
# Now try different settings to see how bias changes!
# Here we'll use a small number of iterations (200) to
# speed things up, but in general bigger is better
# Should be unbiased
iterate(2000, 0, 0, 200)
# Should still be unbiased
iterate(2000, 0, 1, 200)
# How much bias?
iterate(2000, 1, 1, 200)
# Now?
iterate(2000, .1, .1, 200)
# Does it make a difference whether the effect
# is stronger on X or Y?
iterate(2000, .5, .1, 200)
iterate(2000, .1, .5, 200)What can we learn from this simulation? First, as we’d expect, with either \(effectWX\) or \(effectWY\) set to zero, there’s no bias - the mean of the estimate minus the truth of 5 is just about zero. We also see that if \(effectWX\) and \(effectWY\) are both 1, the bias is about .5 - half a standard deviation of the error term - and about 50 times bigger than if \(effectWX\) and \(effectWY\) are both .1. Does the size of the bias have something to do with the product of \(effectWX\) and \(effectWY\)? Sorta seems that way… it’s a question you could do more simulations about!
15.4 Power Analysis with Simulation
15.4.1 What Is Power Analysis?
Statistics is an area where the lessons of children’s television are more or less true: if you try hard enough, anything is possible. It’s also an area where the lessons of violent video games are more or less true: if you want to solve a really tough problem, you need to bring a whole lot of firepower (plus, a faster computer can really help matters).
Power analysis. An analysis that links the sample size, effect size, and the statistical precision of an estimate, often with a goal of figuring out whether the sample size is big enough that, if your hypothesized effect size is accurate, you’re likely to reject the null.
Power analysis is the process of trying to figure out if the amount of firepower you have is enough. While there are calculators and analytical methods out there for performing power analysis, power analysis is very commonly done by simulation, especially for non-experimental studies.425 Power analysis calculators have to be specific to your estimate and research design. This is feasible in experiments where a simple “we randomized the sample into two groups and measured an outcome” design covers thousands of cases. But for observational data where you’re working with dozens of variables, some unmeasured, it’s infeasible for the calculator-maker to guess your intentions ahead of time. That said, calculators work well for some causal inference designs too, and I discuss one in Chapter 20 that I recommend for regression discontinuity.
Why is it a good idea to do power analysis? Once we have our study design down, there are a number of things that can turn statistical analysis into a fairly weak tool and make us less likely to find the truth:
- Really tiny effects are really hard to find - good luck seeing an electron without a super-powered microscope! (in \(Y = \beta_1X + \varepsilon\), \(\beta_1\) is tiny)
- Most statistical analyses are about looking at variation. If there’s little variation in the data, we won’t have much to go on (\(var(X)\) is tiny)
- If there’s a lot of stuff going on other than the effect we’re looking for, it will be hard to pull the signal from the noise (\(var(\varepsilon)\) is big)
- If we have really high standards for what counts as evidence, then a lot of good evidence is going to be ignored, so we need more evidence to make up for it (if you’re insisting on a really tiny \(se(\hat{\beta}_1)\))
Conveniently, all of these problems can be solved by increasing our firepower, by which I mean sample size. Power analysis is our way of figuring out exactly how much firepower we need to bring. If it’s more than we’re willing to provide, we might want to turn around and go back home.
If you have bad power and can’t fix it, you might think about doing a different study. The downsides of bad statistical power aren’t just “we might do the study and not get a significant effect - oh darn.” Bad power also makes it more likely that, if you do find a statistical effect, it’s overinflated or not real! That’s because with bad power we basically only reject the null as a fluke. Think about it this way - if you want to reject the null at the 95% level that a coin flip is 50/50 heads/tails, you need to see an outcome that’s less than 5% likely to occur with a fair coin. If you flip six coins (tiny sample, bad power), that can only happen with all-heads or all-tails. Even if the truth is 55% heads so the null really is false, the only way we can reject 50% heads is if we estimate 0% heads or 100% heads. Both of those are actually way further from the truth than the 50% we just rejected! This is why, when you read about a study that seems to find an enormous effect of some innocuous thing, it often has a tiny sample size (and doesn’t replicate).
Power analysis can be a great idea no matter what kind of study you’re running. However, it’s especially helpful in three cases:
- If you’re looking for an effect that you think is probably not that central or important to what’s going on - it’s a small effect, or a part of a system where a lot of other stuff is going on (\(\beta_1\) times \(var(X)\) is tiny relative to \(var(Y)\)) a power analysis can be a great idea - the sample size required to learn something useful about a small effect is often much bigger than you expect, and it’s good to learn that now rather than after you’ve already done all the work
- If you’re looking for how an effect varies across groups (in regression, if you’re mostly concerned with the interaction term), then a power analysis is a good idea. Finding differences between groups in an effect takes a lot more firepower to find than the effect itself, and you want to be sure you have enough
- If you’re doing a randomized experiment, a power analysis is a must - you can actually control the sample size, so you may as well figure out what you need before you commit to way too little!
15.4.2 What Power Analysis Does
Power analysis balances five things, like some sort of spinning-plate circus act. Using \(X\) to represent treatment and \(Y\) for the outcome, those things are:
- The true effect size (coefficient in a regression, a correlation, etc.)
- The amount of variation in the treatment (\(var(X)\))
- The amount of other variation in \(Y\) (either the variation from the residual after explaining \(Y\) with \(X\), or just \(var(Y)\))
- Statistical precision (the standard error of the estimate or statistical power, i.e., the true-positive rate)
- The sample size
A power analysis holds four of these constant and tells you what the fifth can be. So, for example, it might say “if we think the effect is probably A, and there’s B variation in \(X\), and there’s C variation in \(Y\) unrelated to \(X\), and you want to have statistical precision of D or better, then you’ll need a sample size of at least E.” This tells us the minimum sample size necessary to get sufficient statistical power.
Minimum sample size. The smallest sample size that produces a certain statistical precision given a certain effect size, treatment variation, and unrelated outcome variation.
Minimum detectable effect. The smallest effect size that produces a certain statistical precision given a certain sample size, treatment variation, and unrelated outcome variation.
Or we can go in other directions. “If you’re going to have a sample size of A, and there’s B variation in \(X\), and there’s C variation in \(Y\) unrelated to \(X\), and you want to have statistical precision of D or better, then the effect must be at least as large as E.” This tells us the minimum detectable effect, i.e., the smallest effect you can hope to have a chance of reasonably measuring given your sample size.
You could go for the minimum acceptable any one of the five elements. However, minimum sample size and minimum detectable effect are the most common ones people go for, followed closely by the level of statistical precision.
Statistical power, or the true-positive rate. The probability of rejecting the null hypothesis, when the null hypothesis is actually false.
How about that “statistical precision” thing? What do I mean by that, specifically? Usually, statistical precision in power analysis is measured by a target level of statistical power (thus the name “power analysis”). Statistical power, also known as the true-positive rate, is a concept specific to hypothesis testing. Consider a case where we know the null hypothesis is false. If we had an infinitely-sized sample, we’d definitely reject that false null. However, we don’t have an infinite sample. So, due to sampling variation, we’ll correctly reject the null sometimes, and falsely fail to reject it other times. The proportion of times we correctly reject the false null is the true-positive rate, also known as statistical power. If we reject a false null 73% of the time, that’s the same as saying we have 73% power to reject that null.426 Power is specific to the null we’re trying to reject. It’s pretty easy to reject a null that’s completely ridiculous, but rejecting a null that’s near the truth, but is not the truth, takes a lot more firepower.
Statistical power is dependent on the kind of test you’re running, too - if you are doing a hypothesis test at the 95% confidence level, you’re more likely to reject the null (and thus will have higher statistical power) than if you’re doing a hypothesis test at the 99% confidence level. The more careful you are about false positives, the more likely you are to get a false negative. So there’s a tradeoff there.
Power analyses don’t have to be run with statistical power in mind. In fact, you don’t necessarily need to think about things in terms of “the ability to reject the null,” which is what statistical power is all about. You could run your power analysis with any sort of statistical precision as a goal, like standard errors. Given A, B, and C, what sample size D do you need to make your standard errors E or smaller?
15.4.3 Where Do Those Numbers Come From?
In order to do power analysis, you need to be able to fill in the values for four of those five pieces, so that power analysis can tell you the fifth one. How do we know those things?
We have to make an informed guess. We can use previous research to try to get a sense of how big the effect is likely to be, or how much variation there’s likely to be in X. If there’s no prior research, do what you can - think about what is likely to be true, make educated guesses. Power analysis at a certain level requires some guesswork.
Other pieces aren’t about guesses but about standards. How much statistical power do you want? The higher your statistical power, the less chance there is of missing a true effect. But that means your sample size needs to go up a lot. Often people use a rule of thumb here. In the past, a goal of 80% statistical power has been standard. These days I see 90% a lot more often.
In practical terms, power analysis isn’t a super-precise calculation; it’s guidance. It doesn’t need to be exact. A little guesswork (although ideally as little as possible) is necessary. After all, even getting the minimum sample size necessary doesn’t guarantee your analysis is good, it just gives you a pretty good chance of finding a result if it’s there. Often, people take the results of their power analysis as a baseline and then make sure to overshoot the mark, under the assumption they’ve been too conservative. So don’t worry about being fully accurate, just try to make the numbers in the analysis close enough to be useful.
15.4.4 Coding a Power Analysis Simulation
Most of the pieces for doing a power analysis simulation we already have in place from the rest of the chapter. The “Breaking Things” section is the most informative, since many power analysis simulations will want to try things while changing quite a few settings.
Let’s consider three components we’ve already gone over:
Data creation. We need a data creation function that mimics what we think the actual data generating process looks like. That’s right, it’s back to drawing out the causal diagram of our research setting! You already did this, right?
You can simplify a bit - maybe instead of sixteen variables you plan to measure and control for, and eight you don’t, you can combine them all into “controls” and “unmeasured variables.” But overall you should be thinking about how to create data as much like the real world as possible. You’ll also want to try to match things like how much variation there is - how much variation in treatment do you expect there to be?427 If you have observational data, maybe you can estimate this, and other features of the data, using old data from before your study setting. How much variation in the outcome? In the other variables? You won’t get it exactly, but get as close as you can, even if it’s only an educated guess.
Estimation and storing the results. Once we have our data, we need to perform the analysis we plan to perform. This works the exact same way it did before - just perform the analysis you plan to perform in your study, but using the simulated data!
The only twist comes if you are planning to use statistical power as your measure of statistical precision. How can we pull statistical power out of our analysis?
Remember, statistical power is the proportion of results that are statistically significant (assuming the null is false, which we know it is as long as our data creation process has a nonzero effect). So in each iteration of the simulation, we need to pull out a binary variable indicating whether the effect was statistically significant and return that. Then, the proportion of results that are significant across all the simulations is your statistical power. 45% of iterations had a significant effect (and your null was false)? 45% statistical power!
Getting out whether an effect was significant is a touch harder than pulling out the coefficient or standard error. In R, for a model m you can load the broom package and do d <- tidy(m). Then, d$p.value[2] < .05 will be TRUE if the p-value is below .05 (for a 95% significance test). The [2] there assumes your coefficient of interest is the second one (take a look at the tidy() output for one of the iterations to find the right spot for your coefficient).
In Stata, you’ll need to dip a bit into the Mata matrix algebra underlying many of Stata’s commands. Don’t worry - it won’t bite. Following a regression, pull out the matrix of results with matrix R = r(table). Then, get the p-value out with matrix p = R["pvalue","X"]. Then, you can put that p-value where you want by referring to it with p[1,1].
In Python, mercifully, things are finally easy. For a model m from statsmodels, we can get the p-value for the coefficient on \(X\) with m.pvalues['X']. Compare that to, say, .05 for a significance test. m.pvalues['X'] < .05 is True if the coefficient is significant at the 95% level, and False otherwise.
Iteration. Running the simulation over multiple iterations works the same as it normally would! No changes there.
However, we might want to add another iteration surrounding that one!428 Who iterates the iterators? Say, for example, we want to know the minimum detectable effect that gives 90% power. How can we find it? Well, we might try a bunch of different effect sizes, increasing them a little at a time until we find one that produces 90% power. “Trying a bunch of different effect sizes” is a job for a loop.
Similarly, if we want to find the minimum necessary sample size to get 90% power, we’d try a bunch of different sample sizes until power reached 90%.
So let’s do the thing. I mentioned earlier that one case where there’s poor power is where we want to know the difference in the effect between two groups, in other words the coefficient on an interaction term in a regression (where one of the terms being interacted is binary).
Let’s see how bad the power actually is! Let’s imagine that we want to know the effect of a reading-training program on children’s reading test scores. Also, we’re interested in whether the effect differs by gender.
In the study we plan to do, we know that the test score is explicitly designed to be normally distributed (many tests are designed with this in mind) with a standard deviation of 10.429 I’m about to use this 10 as a reason for creating the test score by adding up all our other determinants and then adding on a normally-distributed error with a standard deviation of 9. Why 9 and not 10? Because the other components we add up to make the test score will increase the standard deviation. This won’t hit a standard deviation of 10 exactly, but it will be close enough.
Other stuff matters for test scores too - let’s pack a bunch of stuff together into a single variable we’ll include as a control \(W\) - a one-standard-deviation change in this control increases test scores by 4.
The reading-training program, in truth, increases test scores by 2 for boys and 2.8 for girls. That’s a pretty big difference - the program is 40% more effective for girls! The training program is administered in a nonrandom way, with higher \(W\) values making it less likely you get the training.
We can estimate the difference in the effect of training between boys and girls with a regression
where we will focus on \(\hat{\beta}_3\).
We want to know the minimum sample size necessary to get 90% power to detect the difference in the effect between boys and girls. Then, let’s imagine we don’t know the true difference in effects, but do know that there are only 2,000 children available to sample, and ask what the smallest possible difference in effects is that we could detect with 90% power with that sample of 2,000. Let’s code!
R Code
library(tidyverse); library(purrr); library(broom)
set.seed(1000)
# Follow the description in the text for data creation. Since we want
# to get minimum sample size and minimum detectable effect, allow both
# sample size and effect to vary.
# diff is the difference in effects between boys and girls
create_data <- function(N, effect, diff) {
d <- tibble(W = rnorm(N),
girl = sample(0:1, N, replace = TRUE)) %>%
# A one-SD change in W makes treatment 10% less likely
mutate(Training = runif(N) + .1*W < .5) %>%
mutate(Test = effect*Training + diff*girl*Training +
4*W + rnorm(N, sd = 9))
return(d)
}
# Our estimation function
est_model <- function(N, effect, diff) {
d <- create_data(N, effect, diff)
# Our model
m <- lm(Test~girl*Training + W, data = d)
tidied <- tidy(m)
# By looking we can spot that the interaction
# term is in the 5th row
sig <- tidied$p.value[5] < .05
return(sig)
}
# Iteration function!
iterate <- function(N, effect, diff, iters) {
results <- 1:iters %>%
map_dbl(function(x) {
# To keep track of progress
if (x %% 100 == 0) {print(x)}
# Run our model and return the result
return(est_model(N, effect, diff))
})
# We want to know statistical power,
# i.e., the proportion of significant results
return(mean(results))
}
# Let's find the minimum sample size
mss <- tibble(N = c(10000, 15000, 20000, 25000))
mss$power <- c(10000, 15000, 20000, 25000) %>%
# Before we had to do function(x) here, but now the argument we're
# passing is the first argument of iterate() so we don't need it
map_dbl(iterate, effect = 2, diff = .8, iter = 500)
# Look for the first N with power above 90%
mss
# Now for the minimum detectable effect
mde <- tibble(effect = c(.8, 1.6, 2.4, 3.2))
mde$power <- c(.8, 1.6, 2.4, 3.2) %>%
map_dbl(function(x) iterate(N = 2000, effect = 2,
diff = x, iter = 500))
# Look for the first effect wth power above 90%
mdeStata Code
set seed 1000
* Follow the description in the text for data creation. Since we want to
* get minimum sample size and minimum detectable effect, allow both
* sample size and effect to vary.
* diff is the difference in effects between boys and girls
capture program drop create_data
program define create_data
local N = `1'
local effect = `2'
local diff = `3'
clear
set obs `N'
g W = rnormal()
g girl = runiformint(0,1)
* A one-SD change in W makes treatment 10% less likely
g Training = runiform() + .1*W < .5
g Test = `effect'*Training + `diff'*girl*Training + ///
4*W + rnormal(0,9)
end
* Our estimation function
capture program drop est_model
program define est_model
local N = `1'
local effect = `2'
local diff = `3'
create_data `N' `effect' `diff'
* Out model
reg Test i.girl##i.Training W
end
* Iteration function!
capture program drop iterate
program define iterate
local N = `1'
local effect = `2'
local diff = `3'
local iters = `4'
quietly {
* Parent data
clear
set obs `iters'
g significant = .
forvalues i = 1(1)`iters' {
* Keep track of progress
if (mod(`i',100) == 0) {
display `i'
}
preserve
est_model `N' `effect' `diff'
restore
* Get significance of the interaction (1.girl#1.Training)
matrix R = r(table)
matrix p = R["pvalue","1.girl#1.Training"]
local sig = p[1,1] < .05
replace significant = `sig' in `i'
}
* Get proportion significant
summ significant
}
display r(mean)
end
* Let's find the minimum sample size
* Look for the first N with power above 90%
foreach N in 10000 15000 20000 25000 {
display "Sample size `N'. Power:"
iterate `N' 2 .8 500
}
* Now for the minimum detectable effect
* Look for the first effect with power above 90%
foreach diff in .8 1.6 2.4 3.2 {
display "Effect difference `diff'. Power:"
iterate 2000 2 `diff' 500
}Python Code
import pandas as pd
import numpy as np
import statsmodels.formula.api as smf
rng = np.random.default_rng(1000)
# Follow the description in the text for data creation. Since we want to
# get minimum sample size and minimum detectable effect, allow both sample
# size and effect to vary.
# diff is the difference in effects between boys and girls
def create_data(N, effect, diff):
d = pd.DataFrame({'W': rng.normal(size = N),
'girl': rng.choice(2, size = N)})
# A one-SD change in W makes treatment 10% less likely
d['Training'] = rng.uniform(size = N) + .1*d['W'] < .5
d['Test'] = effect*d['Training'] + diff*d['girl']*d['Training']
d['Test'] = d['Test'] + 4*d['W'] + rng.normal(scale = 9, size = N)
return(d)
# Our estimation function
def est_model(N, effect, diff):
d = create_data(N, effect, diff)
# Our model
m = smf.ols('Test~girl*Training + W', data = d).fit()
# By looking we can spot that the name of the
# interaction term is girl:Training[T.True]
sig = m.pvalues['girl:Training[T.True]'] < .05
return(sig)
# Iteration function!
def iterate(N, effect, diff, iters):
results = [est_model(N, effect, diff) for i in range(0,iters)]
# We want to know statistical power,
# i.e., the proportion of significant results
return(np.mean(results))
# Let's find the minimum sample size
mss = [[N, iterate(N, 2, .8, 500)] for
N in [10000, 15000, 20000, 25000]]
# Look for the first N with power above 90%
pd.DataFrame(mss, columns = ['N','Power'])
# Now for the minimum detectable effect
mde = [[diff, iterate(2000, 2, diff, 500)] for
diff in [.8, 1.6, 2.4, 3.2]]
pd.DataFrame(mde, columns = ['Effect', 'Power'])What do we get from these analyses? From the minimum-sample-size power analysis we get Table 15.1. In that table, we see that we find 88.2% power with a sample size of 20,000, and 93.8% power with a sample size of 25,000. So our sweet spot of 90% power must be somewhere between them. We could try increasing the number of iterations, or trying different \(N\)s between 20,000 and 25,000 to get a more precise answer, but “a little more than 20,000 observations is necessary to get 90% power on that interaction term” is pretty good information as is.430 And if all of them had 90% power (or none of them did) that would be an indication we’d need to try smaller (larger) sample sizes. That’s how I found the range of sample sizes to iterate over here.
Table 15.1: Results of Minimum Sample Size Power Analysis Simulation
| N | Power |
|---|---|
| 10,000 | 0.602 |
| 15,000 | 0.788 |
| 20,000 | 0.882 |
| 25,000 | 0.938 |
Wait, uh, twenty thousand? For that massive interaction term (how often do you think a treatment intended to apply to everyone is really 40% more effective for one group than another)? Yep! “40% more effective” isn’t as important as “.8 relative to the standard deviation of 10 for \(Test\).”
It’s no surprise that if we have only 2,000 observations, we’re nowhere near 90% power for the true interaction effect of .8. But what’s the minimum detectable effect we can detect with our sample size of 2,000? The results are in Table 15.2.
Table 15.2: Results of Minimum Detectable Effect Power Analysis Simulation
| Effect | Power |
|---|---|
| 0.8 | 0.164 |
| 1.6 | 0.528 |
| 2.4 | 0.838 |
| 3.2 | 0.976 |
In Table 15.2 we reach 90% power with an effect somewhere between 2.4 and 3.2. Again, we could try with more iterations and more effect sizes between 2.4 and 3.2 to get more precise, but this gives us a ballpark.
If we only have 2,000 observations to work with, we’d need the effect for girls to be something like 220% of the effect for boys (or 40% of the effect but with an opposite sign) to be able to detect gender differences in the effect with 90% power. Do we think that’s a reasonable assumption to make? If not, maybe we shouldn’t estimate gender differences in the effect for this study, even if we really want to. That’s what power analysis is good at - knowing when a fight is lost and you should really go home.431 There are some statistical issues inherent to interactions that worsen their power, but the real problem is that interaction terms are in truth often small. Treatment effects vary across groups, but that variation is often much smaller than the effect itself. So however many observations we need to find the main effect, we need many more to find those tiny interactions. Small effects are very hard to find with statistics!
15.5 Simulation with Existing Data: The Bootstrap
Seriously, the third chapter in a row where I’m talking about the bootstrap? Well, deal with it. The bootstrap is basically magic. Don’t you want magic in your life?432 The imaginary person I’m responding to here is such an ingrate.
Bootstrapping is the process of repeating an analysis many times after resampling with replacement. Resampling with replacement means that, if you start with a data set with \(N\) observations, you create a new simulated data set by picking \(N\) random observations from that data set… but each time you draw an observation, you put it back so you might draw it again. If your original data set was 1, 2, 3, 4, then a bootstrap data set might be 3, 1, 1, 4, and another one might be 2, 4, 1, 4. Alternately, you could create random sample weights from 0 to 1 instead of making each observation a stark in/out decision, although we won’t code this up ourselves here (Rubin 1981Rubin, Donald B. 1981. “The Bayesian Bootstrap.” The Annals of Statistics, 130–34.).
The resampling process mimics the sampling distribution in the actual data. On average, the means of the variables will match your means, as will the variances, the correlations between the variables, and so on. In this way, we can run a simulation using actually-existing data. The resampling-with-replacement process replaces the create_data function in our simulation. One of the toughest parts of doing a simulation is figuring out how to create data that looks like the real world. If we can just use real-world data, that’s miles better.
The only downside is that, unlike with a simulation where you create the data yourself, you don’t know the true data generating process. So you can’t use bootstrapping to, say, check for bias in your estimate. You can see what your bootstrap-simulated estimate is, but you don’t know the truth to compare it to (since you didn’t create the data yourself), so you can’t check whether you’re right on average.
This means that the bootstrap, as a simulation tool, is limited to cases where the question doesn’t rely on comparing your results to the truth. Usually this means using the bootstrap to estimate standard errors, since by mimicking the whole resampling process, you automatically simulate the strange interdependencies between the variables, allowing any oddities in the true sampling distribution to creep into your bootstrap-simulated sampling distribution.
Standard errors aren’t all it’s good for, though! Machine learning people use bootstrapping for all sorts of stuff, including using it to generate lots of training data sets for their models. You can also use a bootstrap to look at the variation of your model more generally. For example, if you make a bootstrap sample, estimate a model, generate predicted values from that model, and then repeat, you can see how your predictions for each observation vary based on the sampling variation of the whole model, or just aggregate all the simulated predictions together to get your actual prediction.433 This process has the name “bagging” which on one hand I like as a term, but on the other hand really sounds like some super-dorky-in-retrospect dance craze from, like, 1991. \(^,\)434 One important note is that if you’re planning to use your bootstrap distribution to do a hypothesis test, you want to first shift the distribution so it has the unbootstrapped estimate as the mean, before you compare the distribution to the null. This produces the appropriate comparison you’re going for with a hypothesis test.
How can you do a bootstrap? Borrowing from Chapter 13:
- Start with a data set with \(N\) observations.
- Randomly sample \(N\) observations from that data set, allowing the same observation to be drawn more than once. This is a “bootstrap sample”.
- Estimate our statistic of interest using the bootstrap sample.
- Repeat steps 1-3 a whole bunch of times.
- Look at the distribution of estimates from across all the times we repeated steps 1-3.435 If your data or estimate follows a super-weird distribution, perhaps one with an undefined mean (see Chapter 23), you’ll want to drop some of the tails of your bootstrap distribution before using it. The standard deviation of that distribution is the standard error of the estimate. The 2.5th and 97.5th percentiles show the 95% confidence interval, and so on.
How many times is “a bunch of times” in step 4? It varies. When you’re just testing out code, you can do it a few hundred times to make sure things work, but you wouldn’t trust those actual results. Really, you should do it as many times as you feasibly can given the computing power available to you. Be prepared to wait! For anything serious, you probably want to do it at least a few thousand times.
Bootstrapping comes with its own set of assumptions to provide good estimates of the standard error. We need a reasonably large sample size. We need to be sure that our data is at least reasonably well behaved - if the variables follow extremely highly skewed distributions, bootstrap will have a hard time with that. We also need to be careful in accounting for how the observations might be related to each other. For example, if there are individuals observed multiple times (panel data), we may want a “cluster bootstrap” that samples individuals with replacement, rather than observations (getting the 2005 and 2007 observations for you, and the 2005 observation for me twice, might not make sense). Or if the error terms are correlated with each other - clustered, or autocorrelated - the bootstrap’s resampling approach needs to be very careful to deal with it.
15.5.1 Coding a Bootstrap
In the interest of not repeating myself too much, this section will be short. There are already coding examples for using built-in bootstrap standard error tools in Chapter 13 on Regression, and for inverse probability weighting with regression adjustment in Chapter 14 on Matching.
What’s left to do? In this section, I’ll show you how to do two things:
- A generic demonstration of how to perform a bootstrap “by hand.” The example will provide bootstrap standard errors. For R and Stata, bootstrap standard errors are easier done by the built-in methods available for most models, but the code here gives you a better sense of what’s happening under the hood, can be easily manipulated to do something besides standard errors, which those built-in methods can’t help with, and can work with commands that don’t have built-in bootstrap standard error methods.
- After this short refresher, I’ll go into a little more about some variants of the bootstrap
Let’s do a bootstrap by hand! The process here is super simple. We just copy all the stuff we have already been doing with our simulation code. Then, we replace the create_data function, which created our simulated data, with a function that resamples the data with replacement.
Or, we can save ourselves a little work and skip making the bootstrap process its own function - usually it only takes one line anyway. If we have \(N\) observations, just create a vector that’s \(N\) long, where each element is a random integer from 1 to \(N\) (or 0 to \(N-1\) in Python). Then use that vector to pick which observations to retain. In R and Python, this is simply:
R Code
library(tidyverse)
# Get our data set
data(mtcars)
# Resampling function
create_boot <- function(d) {
d <- d %>%
slice_sample(n = nrow(d), replace = TRUE)
return(d)
# Or more "by-hand"
# index <- sample(1:nrow(d), nrow(d), replace = TRUE)
# d <- d[index,]
}
# Get a bootstrap sample
create_boot(mtcars)Python Code
Easy! You can slot that right into your simulations as normal.
However, when it comes to bootstrapping “by hand” we might want to take a half-measure - doing the estimation method, and returning the appropriate estimate, by hand, but letting a function do the bootstrap sampling and iteration work. All three languages have built-in bootstrap-running commands. We already saw Stata’s in Chapter 14.
For all of these, we need to specify an estimation function first. Unlike in our previous code examples, that estimation function won’t call a data-creation function to get data to estimate with. Instead, we’ll call that estimation function with our bootstrapping function, which will provide a bootstrapped sample to be estimated, take the estimates provided, and aggregate them all together.
R Code
library(tidyverse); library(boot)
set.seed(1000)
# Get our data set
data(mtcars)
# Our estimation function. The first argument must be the data
# and the second the indices defining the bootstrap sample
est_model <- function(d, index) {
# Run our model
# Use index to pick the bootstrap sample
m <- lm(hp ~ mpg + wt, data = d[index,])
# Return the estimate(s) we want to bootstrap
return(c(coef(m)['mpg'],
coef(m)['wt']))
}
# Now the bootstrap call!
results <- boot(mtcars, # data
est_model, # Estimation function,
1000) # Number of bootstrap simulations
# See the coefficient estimates and their bootstrap standard errors
resultsStata Code
set seed 1000
* Get data
sysuse auto.dta, clear
* Estimation model
capture program drop est_model
program def est_model
quietly: reg price mpg weight
end
* The bootstrap command is only a slight difference from the simulate and
* bstat approach in the Matching chapter. Just expanding the toolkit!
bootstrap, reps(1000): est_modelPython Code
import pandas as pd
import numpy as np
from resample.bootstrap import bootstrap
import statsmodels.formula.api as smf
import seaborn as sns
rng = np.random.default_rng(1000)
# Get our data to bootstrap
iris = sns.load_dataset('iris')
# Estimation - the first argument should be for the bootstrapped data
def est_model(d):
# bootstrap() makes an array, not a DataFrame
d = pd.DataFrame(d)
# Oh also it tossed out the column names
d.columns = ['sepal_length','sepal_width','petal_length']
m = smf.ols(formula = 'sepal_length ~ sepal_width + petal_length',
data = d).fit()
coefs = [m.params['sepal_width'], m.params['petal_length']]
return(coefs)
# Bootstrap the iris data, estimate with est_model, and do it 1000 times
# We toss out the string variable; this tends to confuse bootstrap
b = bootstrap(sample = iris[['sepal_length','sepal_width','petal_length']],
fn = est_model, size = 1000)
# Get our standard errors
bDF = pd.DataFrame(b, columns = ['SW','PL'])
bDF.std()Not too bad! Okay, not actually easier so far than doing it by hand, but there’s a real benefit here, in that these purpose-built bootstrap commands often come with options that let you bootstrap in ways that are a bit more complex than simply sampling each observation with replacement. Or they can provide ways of analyzing the distribution of outputs in slightly more sophisticated ways than just taking the mean and standard deviation.
One of the nicest benefits you get from these commands is the opportunity to do some bias correction. Here bias doesn’t refer to failing to identify our effect of interest, but rather how the shape of the data can bias our statistical estimates. Remember how I mentioned that bootstrap methods can be sort of sensitive to skewed data? Skewed data will lead to skewed sampling distributions, which will affect how you get an estimate and standard error from that sampling distribution.
But there are ways to correct this bias, the most common of which is “BCa confidence intervals.” This method looks at the proportion of times that a bootstrap estimate is lower than the non-bootstrapped estimate (if the distribution isn’t skewed, this should be 50%), and the skew of the distribution.436 They also use a cousin of bootstrapping and (from Chapter 13) cross-validation called the “jackknife,” in which for a sample with \(N\) observations the simulation is run \(N\) times, each time omitting exactly one observation, so each observation is omitted from one sample. This is used here to see how influential each observation is on the estimates. It can also be used to generate out-of-sample predictions for the currently-excluded observation. Then, the BCa confidence intervals you get account for the skewing of the bootstrap sampling distribution.437 Interestingly, they’re not symmetric around the estimate - if, for example, skew means there are “too many” bootstrap estimates that are lower than (to the left of) the non-bootstrapped estimate, then we know a bit more about the left-hand side of the distribution, and the 95% confidence interval doesn’t need to be very far out. But on the right, we need to go farther out. You can then use these confidence intervals for hypothesis testing.
In R, you can pass your results from boot to boot.ci(type = 'bca') to get the BCa confidence intervals. In Stata, add the bca option to your bootstrap command, and then once it’s done do estat bootstrap, bca (although in my experience this process is a bit finicky). In Python, import and run confidence_interval instead of bootstrap from resample.bootstrap.
15.5.2 Bootstrap Variants
Like just about every statistical method, the bootstrap stubbornly refuses to just be one thing, and instead there are a jillion variations on it. The fact that I’ve been referring to the thing I’ve been showing you as “the bootstrap” instead of more precisely as “pairs bootstrap” has almost certainly angered someone. I’m not entirely sure who, though.438 The literature on exactly which format of bootstrap to use, with which resampling method, with which random generation process, with which adjustment, is deep and littered with caveats like “this particular method seemed to do well in this particular instance.” Unfortunately, the learning curve is steep. If you’re really interested in diving in, good luck!
One classic alternative is the Bayesian bootstrap, which is pretty straightforward (Rubin 1981Rubin, Donald B. 1981. “The Bayesian Bootstrap.” The Annals of Statistics, 130–34.). Resampling with replacement is just like assigning sample weights to your data (see 13, where all the weights are integers representing the number of times you sampled that observation. But why limit yourself to integers? Instead of resampling, you could instead draw continuous sample weights from the Dirichlet distribution (rdirichlet() from extraDistr or a few other packages in R or np.random.default_rng.dirichlet() in Python),439 There’s no direct command in Stata, but if you make random Gamma distribution data with rgamma(), then divide by the sum of all the random data you drew, that will give you Dirichlet data. setting the \(\alpha\) parameter to 4 (Shao and Tu 2012Shao, Jun, and Dongsheng Tu. 2012. The Jackknife and Bootstrap. Springer Science & Business Media.). The rest is the same. The result is usually faster to run, and is a little less noisy from iteration to iteration, because the continuous weights are not quite as jarring as the stark in/out decision of the standard bootstrap. You also run into fewer issues where, for example, your model can no longer be estimated because you happened to drop just the wrong set of observations. Perhaps one day it will be more popular than the standard bootstrap, although that day is not today.
That’s just one. There are far more variants of the bootstrap than I’ll go into here, but I will introduce a few more, specifically the ones that focus on non-independent data. Remember when I said that the bootstrap only works if the observations are unrelated to each other? It’s true! Or at least it’s true for the basic version. But what if we have heteroskedasticity? Or autocorrelation? Or if our observations are clustered together? It’s not the case that the bootstrap can’t work, but it is the case that our bootstrapping procedure has to account for the relationship between the observations.
The cluster/block bootstrap seems like a good place to start, since I’ve already talked about it a bit. The cluster bootstrap is just like the regular bootstrap, except instead of resampling individual observations, you instead resample clusters of observations, also known as blocks. That’s the only difference!
You’d want to think about using a cluster bootstrap any time you have hierarchical or panel data.440 In these cases, it’s called “cluster bootstrap” instead of “block bootstrap” by convention. Hierarchical data is when observations are nested within groups. For example, you might have data from student test scores. Those students are within classrooms, which are within schools, which are within regions, which are within countries.
Depending on the analysis you’re doing, that might not necessitate a cluster bootstrap (after all, all data is hierarchical in some way). But if the hierarchical structure is built into your model - say, you are incorporating classroom-level characteristics into your model, then an individual-observation bootstrap might not make much sense.
Not all hierarchical data sets call for a cluster bootstrap, but when it comes to panel data, it’s often what you want. In panel data we observe the same individual (person, firm, etc.) multiple times. We’ll talk about this a lot more in Chapter 16. When this happens, it doesn’t make too much sense to resample individual observations, since that would sort of split each person in two and resample only part of them. So we’d want to sample individuals, and all of the observations we have for that individual, rather than each observation separately.
Another place where the cluster/block bootstrap pops up is in the context of time series data.441 This time it’s “block bootstrap” instead of “cluster bootstrap” by convention. Go figure. In time series data, the same value is measured across many time periods. This often means there is autocorrelation. When data is correlated across time, if you just resample randomly you’ll break those correlation structures. Also, if your model includes lagged values, you’re in trouble if you happen not to resample the value just before yours!
So, when applying a bootstrap to time series data, the time series is first divided into blocks of continuous time. The blocks themselves are often determined by one of several different available optimal-block-length algorithms (Politis and White 2004Politis, Dimitris N., and Halbert White. 2004. “Automatic Block-Length Selection for the Dependent Bootstrap.” Econometric Reviews 23 (1): 53–70.). If you have daily stock index prices from 2000 through 2010, maybe each month ends up its own block. Or every six weeks is a new block, or every year, or something, or maybe some blocks are longer than others. The important thing is that within each block you have a continuous time series. Then, you resample the blocks instead of individual observations.442 A common alternative to this is the “moving block bootstrap,” which sort of has a sliding window that gets resampled instead of splitting the data up into fixed chunks.
The cluster/block bootstrap can be implemented in Stata by just adding a cluster() option onto your regular bootstrap command. In R, you can use the cluster option in the vcovBS() function in the sandwich package if what you want is bootstrap standard errors on a regression model (pass the vcovBS() result to the vcov argument in your modelsummary table). There doesn’t appear to be a more general implementation in R or Python that works with any function like bootstrap does. There doesn’t appear to be any cluster bootstrap implementation at all in Python, but several varieties of block bootstrap are in arch.bootstrap in the arch package, and there is a wild cluster bootstrap implementation available (later in the chapter).
Thankfully, the cluster bootstrap is not too hard to do by hand in R and Python. Just (1) create a data set that contains only the list of ID values identifying individuals, (2) do a regular bootstrap resampling on that data set, and (3) use a merge to merge that bootstrapped data back to your regular data, using a form of merge that only keeps matches (inner_join() in R, or d.join(how = 'inner') in Python). This will copy each individual the appropriate number of times. This same method could also be used to do a block bootstrap, but only if you’re willing to define the blocks yourself instead of having an algorithm do it (probably better than you can).
R Code
library(tidyverse)
set.seed(1000)
# Example data
d <- tibble(ID = c(1,1,2,2,3,3),
X = 1:6)
# Now, get our data set just of IDs
IDs <- tibble(ID = unique(d$ID))
# Our bootstrap resampling function
create_boot <- function(d, IDs) {
# Resample our ID data
bs_ID <- IDs %>%
sample_n(size = n(), replace = TRUE)
# And our full data
bs_d <- d %>%
inner_join(bs_ID, by = 'ID',
relationship = 'many-to-many')
return(bs_d)
}
# Create a cluster bootstrap data set
create_boot(d, IDs)Python Code
import pandas as pd
import numpy as np
rng = np.random.default_rng(1000)
# Example data
d = pd.DataFrame({'ID': [1,1,2,2,3,3],
'X': [1,2,3,4,5,6]})
# Now, get our data frame just of IDs
IDs = pd.DataFrame({'ID': np.unique(d['ID'])})
# Our bootstrap resampling function
def create_boot(d, IDs):
# Resample our ID data
N = IDs.shape[0]
index = rng.choice(N, size = N)
bs_ID = IDs.iloc[index]
# And our full data
bs_d = d.merge(bs_ID, how = 'inner', on = 'ID')
return(bs_d)
# Create a cluster bootstrap data set
create_boot(d, IDs)Another popular type of bootstrap actually works in a very different way. In residual resampling, instead of resampling whole observations, you instead resample the residuals. Huh?
Under residual resampling, you follow these steps:
- Perform your analysis using the original data.
- Use your analysis to get a predicted outcome value \(\hat{Y}\), and also the residual \(\hat{r}\), which is the difference between the actual outcome and the predicted outcome, \(\hat{r} = Y - \hat{Y}\).
- Perform bootstrap resampling to get resampled residuals \(r_b\).
- Create a new outcome variable by adding the residual to the actual outcome, \(Y_b = Y + r_b\).
- Estimate the model as normal using \(Y_b\).
- Store whatever result you want, and iterate steps 3-5.
The idea here is that the predictors in your model never change with this process. So whatever kind of interdependence they have, you get to keep that! A downside of residual resampling is that it doesn’t perform well if the errors are in any way related to the predictors—this is a stronger assumption than even the OLS assumption that errors are on average unrelated to predictors.
Another popular form of the bootstrap in this vein, which both has a very cool name and gets around the unrelated-error assumption, is the wild bootstrap. The wild bootstrap is popular because it works under heteroskedasticity - even when we don’t know the exact form the heteroskedasticity takes. It also performs well when the data is clustered. We already have the cluster/block bootstrap for that. However, the cluster bootstrap doesn’t work very well if the clusters are of very different sizes. If your classroom has 100 kids in it, but mine has five, then resampling classrooms can get weird real fast. The cluster-based version of the wild bootstrap works better in these scenarios. The cluster-based version of the wild bootstrap also works much better than regular sandwich-based clustered standard errors when there are a small number of clusters, where “small” could even be as large as 50.443 Regular clustered standard errors get their results from asymptotic assumptions, i.e., they’re proven to work as intended as the number of clusters gets somewhere near infinity. In practice, for clustered errors, 50 is probably near enough to infinity to be fine. But the wild cluster bootstrap makes no such assumption, and so works much better with smaller numbers of clusters.
Without getting too into the weeds, let’s talk about what the wild bootstrap is. For a wild bootstrap, you follow the general outline of residual resampling. Except you don’t resample the residual. You keep each residual lined up with its original observation each time you create the bootstrap data.
Instead of resampling the residuals, you first transform them according to a transformation function, and then multiply them by a random value with mean 0 (plus some other conditions).444 The mean-zero part means that many of these random values are negative. Some observations get the signs of their residuals flipped. That’s it! You take your random value for each observation, multiply by the transformed residual, use that to get your new outcome value \(Y_b\). Estimate the model with that, then iterate.
Do note that there are quite a few options for which transformation function and which random distribution to draw from. Different choices seem to perform better in different scenarios, so I can’t give any blanket recommendations. If you want to use a wild bootstrap, you might want to do a little digging about how it applies to your particular type of data. If you do want to get into the weeds, I can recommend a nice introduction by Martin (2017Martin, Michael A. 2017. “Wild Bootstrap.” In Wiley StatsRef: Statistics Reference Online, 1–6. John Wiley & Sons, Ltd.).
It seems incomprehensible as to why the wild bootstrap actually works. But the short of it is that the random value you multiply the residual by does manage to mimic the sampling distribution. Plus, it does so in a way that respects the original model, making things a bit more cohesive for the coefficients you’re trying to form sampling distributions for.
Wild bootstrap standard errors can be implemented in R using the type argument of vcovBS() in the sandwich package, or there are some faster options in the fwildclusterboot package. There are different options for type depending on the kind of distribution you want to use to generate your random multipliers, although the default 'wild' option uses the Rademacher distribution, which is simply -1 or 1 with equal probability. In Python there is the wildboottest package with a few different variants of wild cluster bootstraps available.
Page built: 2025-10-17 using R version 4.5.0 (2025-04-11 ucrt)